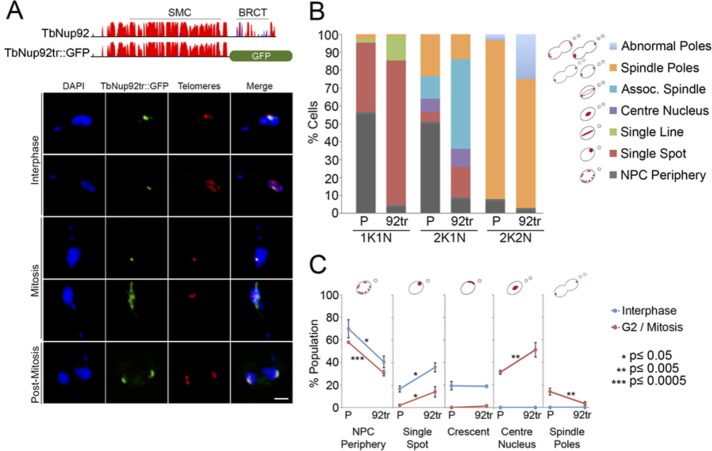
FIGURE 5:
The C-terminal BRCT domain is required for the interphase distribution of TbNup92. (A) Top, predicted secondary structure of TbNup92. The y-axis indicates the confidence score of the predicted secondary structure. Coiled coils are in red and the β-sandwich in blue. TbNup92 contains a central SMC domain and a C-terminal BRCT domain. A single allele of TbNup92 was truncated by replacing residues 639–813 with a GFP tag. The second allele of TbNup92 was replaced with a neomycin resistance gene. Bottom, the nuclear localization of TbNup92 is disrupted in cells expressing TbNup92 lacking the C-terminus containing the BRCT domain (TbNup92tr::GFP, green). Cells were costained with a FISH probe for the telomeres (red). DAPI was used to visualize DNA (blue). Scale bar, 2 μm. (B) The localization of TbNup92::GFP and TbNup92tr::GFP was documented in 100 1K1N, 2K1N, and 2K2N cells. Mean scores of triplicate experimental replicates are shown. Loss of the C-terminus containing the BRCT domain prevents TbNup92 from redistributing to the nuclear periphery and nucleoplasm during interphase. At mitosis, TbNup92tr::GFP is more likely to be distributed along the spindle microtubules, and this coincides with the mislocalization of TbNup92tr::GFP at the spindle poles during late mitosis. (C) A telomere FISH probe was used to visualize the telomeric repeats in TbNup92::GFP- and TbNup92tr::GFP-tagged cells. The localization of the telomeres in 100 interphase and 100 mitotic cells was scored. The mean scores from triplicate experimental replicates are plotted (*p ≤ 0.05; **p ≤ 0.005; ***p ≤ 0.0005). At interphase, the telomeres in TbNup92tr::GFP cells are more likely to be localized as single puncta at the periphery compared with those in TbNup92::GFP cells. In mitotic cells, the telomeres remain fixed at the center of the nucleus for a longer period in TbNup92tr::GFP cells before eventually migrating to the spindle poles.