FIG. 3.
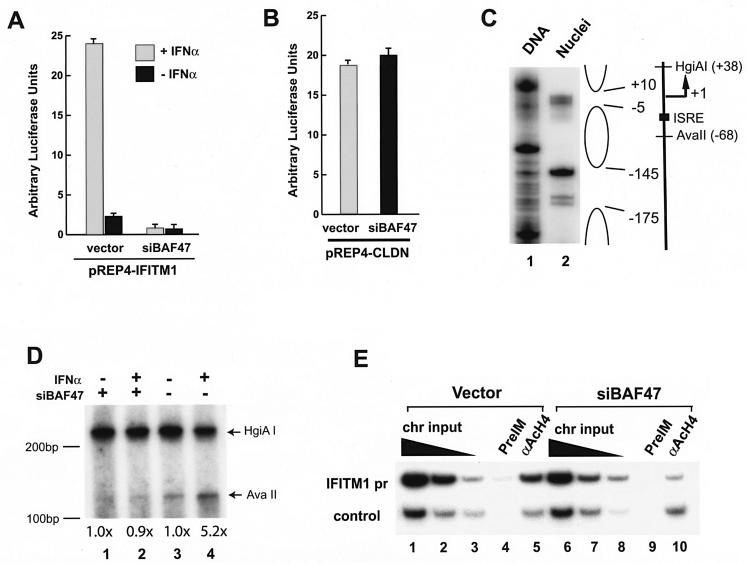
The BAF complex regulates IFITM1 promoter activity by modulating its chromatin structure. (A) Inhibiting the BAF complexes abolished the induction of the IFITM1 promoter by IFN-α. HeLa cells were transfected with the IFITM1 promoter reporter construct, pREP4-IFITM1pr-luc, together with a control vector or the siRNA construct targeting BAF47 (siBAF47) for 48 to 72 h. Following treatment with 500 U of IFN-α/ml for 12 h, the luciferase activity was analyzed with a dual luciferase system from Promega. (B) Knockdown of BAF47 did not inhibit claudin promoter activity. HeLa cells were transfected with the claudin promoter reporter construct and analyzed as described for panel A. (C) The IFITM1 promoter has a positioned nucleosome. The genomic DNA or nuclei isolated from HeLa or SW-13 cells was digested with microccocal nuclease. The double-stranded cleavages in the nucleosomal linker regions were detected by ligation-mediated PCR with primers specific for the IFITM1 promoter sequence (PCR primer −273F [5′ACAGTGAGGTCCTGTACTTGCTGG3′] and labeling primer −261F [5′TGTACTTGCTGGCCTGGGGTG3′]). The open oval on the right indicates the regions protected by the nucleosomal structure. The transcription start site (+1) and direction are indicated. The filled square indicates a potential binding site for ISGF3 (ISRE). The recognition sites for the restriction enzymes (AvaII and HgiAI) are indicated. (D) The BAF complex is required for the chromatin remodeling of the IFITM1 promoter upon IFN-α stimulation. HeLa cells were transfected with control vector or siRNA targeting BAF47 (siBAF47) and selected as described in the legend to Fig. 1. After stimulation with 500 U of IFN-α/ml for 2 h, the cells were permeabilized and briefly digested with AvaII enzyme, followed by complete digestion of the purified genomic DNA with HgiAI enzyme. The cleavage sites were detected by linker ligation-mediated PCR with IFITM1 promoter-specific primers. The data were quantified by PhophorImager analysis. The intensity of the bands produced by AvaII digestion after normalization to the HgiAI digestion is indicated below the panel. The experiments were repeated three times, and similar results were obtained. Size markers are indicated on the left side of the panel. (E) Knocking down BAF47 reduced histone H4 acetylation at the IFITM1 promoter. The HeLa cells were transfected with control vector or siRNA targeting BAF47 (siBAF47) and selected as described in the legend to Fig. 1. The chromatin lysates were prepared and immunoprecipitated with preimmune serum and antibodies against tetra-acetylated histone H4 tail as described previously (24). The IFITM1 promoter sequence and the CSF1 upstream sequence (control) in the immunoprecipitated DNA were analyzed by PCR. The chromatin (chr) input was diluted three times at each step.