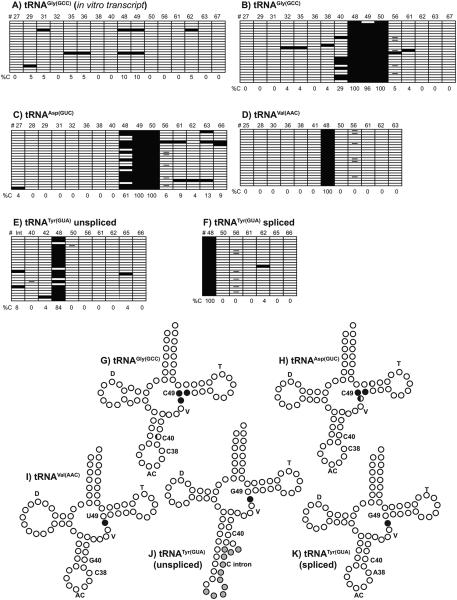
Figure 2. Sodium bisulfite sequencing analysis of procyclic form T. brucei tRNAs.
PCR products from bisulfite treated small RNAs were analyzed by DNA sequencing. A-F) Diagrams of modification patterns of in vitro synthesized tRNAGly(GCC) A) and native T. brucei tRNAs (B-F). Each horizontal row represents one sequencing reaction. Open boxes represent Ts, filled boxes represent Cs, and minus signs (-) represent missing sequencing information in individual clones. Numbers on the top of each diagram represent the position of individual Cs in the tRNA (#), and numbers at the bottom of each diagram represent the percent of sequences with Cs in a particular position (%C). G-K) Patterns of modification with respect to predicted tRNA secondary structures. D is the D-loop, T is the TψC loop, V is the variable region, and AC is the anticodon loop. For unspliced tRNATyr(GUA) (J), gray circles represent bases in the intron. Positions 38, 40, and 49 are labeled for reference. Open circles, cytosines with 10% or less modification (or non-cytosines); ¼ filled circles, cytosines with 11-40% modification; ½ filled circles, cytosines with 41-70% modification; and completely filled circles, cytosines with 71-100% modification.