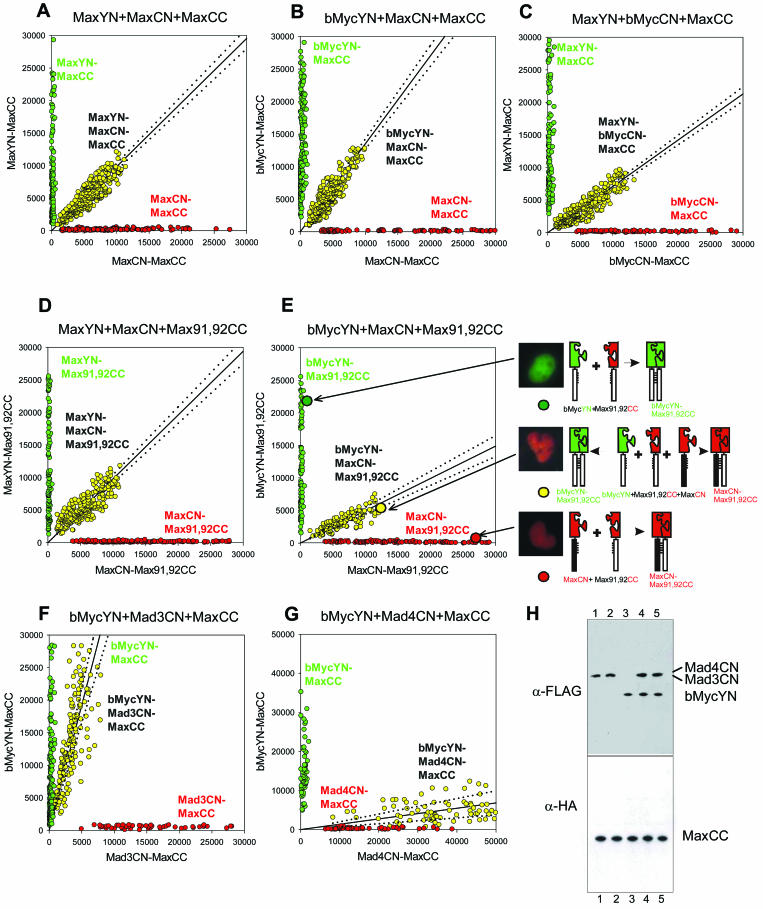
FIG. 7.
Visualization of the competition for dimerization by alternative interaction partners in living cells. (A to G) The proteins indicated above each graph were coexpressed in COS-1 cells in pairs (green and red) and in three-way competition (yellow). The fluorescence emissions corresponding to the spectrally distinct YN-CC and CN-CC complexes were measured in each cell and were plotted for 100 to 250 cells for each combination of proteins. The line shows the linear regression fit to the fluorescence intensities of cells that expressed all three proteins. The 95% confidence intervals for the regression lines are shown as dotted lines. The slope of the linear regression reflects the relative dimerization preferences of the alternative interaction partners used in each experiment. (H) Western blot analysis of the levels of protein expression in the cells whose fluorescence is shown in panels F and G. Plasmids encoding Mad4CN and MaxCC (lane 1), Mad3CN and MaxCC (lane 2), bMycYN and MaxCC (lane 3), Mad4CN, bMycYN, and MaxCC (lane 4), or Mad3CN, bMycYN, and MaxCC (lane 5) were cotransfected into COS-1 cells. Cell lysates were analyzed by Western blot analysis using antibodies that recognize the FLAG (detects bMycYN, Mad3CN, and Mad4CN) or HA (detects MaxCC) epitope (α-FLAG and α-HA, respectively). The lysates were obtained from the same cells that were used to measure the fluorescence intensities shown in panels F and G.