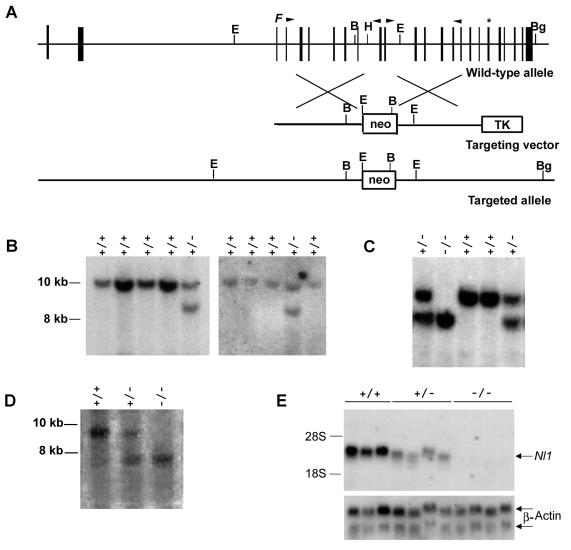
FIG. 1.
Disruption of the Nl1 gene. (A) Schematic representation of the wild-type mouse Nl1 gene, the targeting vector, and the targeted Nl1 allele. Vertical black boxes represent exons. The positions of the neomycin resistance gene (neo) and the herpes simplex virus thymidine kinase gene (TK) are shown. The neo cassette is flanked at its 5′ and 3′ ends by 4.9 and 4.1 kb of Nl1 genomic sequence, respectively. F indicates the position of the convertase cleavage site, and the asterisk shows the position of the NL1 active site. B, BamHI; Bg, BglII; H, HindIII; E, EcoRI. Arrowheads indicate the positions of oligonucleotide primers used to PCR amplify genomic DNA to generate the vector. (B and C) Southern blot analyses of DNA isolated from ES cell clones (B) and mouse tails (C). Genomic DNA was digested with BglII and BamHI and transferred to nylon membranes, and the blots were hybridized with an Nl1 probe located 3′ of the targeting vector sequence (described in Materials and Methods). Fragments from the wild-type (+/+; 9.6 kb) and mutated (−/−; 8.1 kb) alleles were detected. (D) Southern blot analysis of genomic DNA digested with EcoRI, transferred to nylon membranes, and hybridized with an Nl1 probe located 5′ of the targeting vector sequence (described in Materials and Methods). Fragments from the wild-type (+/+; 8.9 kb) and mutated (−/−; 7.3 kb) alleles were detected. (E) Northern blot analysis of total RNA (12.5 μg) isolated from the testes of three wild-type, four Nl1+/−, and four Nl1−/− mice. RNA was hybridized with either the 3′-derived Nl1 or actin probe.