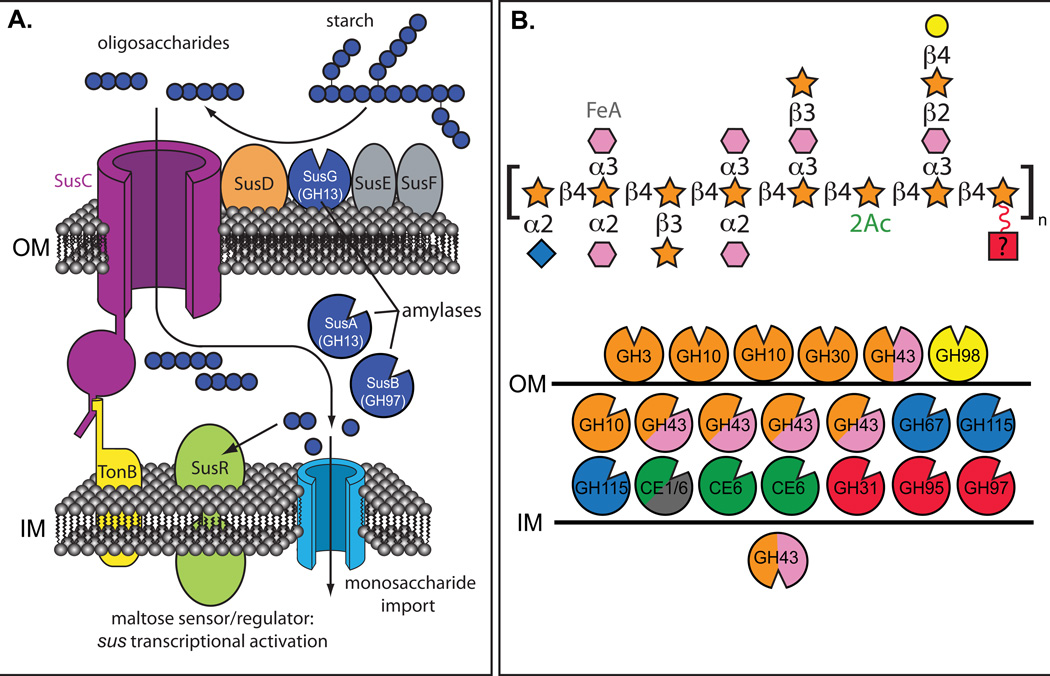
Fig. 2. Variations in functional complexity among Sus-like systems.
Two different representations of Sus-like systems in human gut Bacteroides. A. A model of the B. thetaiotaomicron starch utilization system (Sus). The TonB-dependent transporter SusC works in concert with the starch-binding lipoproteins SusD, SusE, SusF and SusG, which is a glycoside hydrolase family 13 (GH13) α-amylase. Starch binding is initiated by SusD/E/F, followed by initial degradation by SusG, and oligosaccharides are transported into the periplasm via SusC. In the periplasm, maltooligo-saccharides are further degraded to glucose by another GH13 enzyme (SusA, neopullulanse) and a GH97 enzyme (SusB, α-glucosidase). Homologs of the proteins SusC (TonB-dependent porin) and SusD (starch-binding protein) are a hallmark of every Sus-like system, but carbohydrate-binding proteins akin to SusE and SusF as well as glycoside hydrolases, vary substantially between Sus-like loci. B. Depiction of the enzymes encoded in two polysaccharide-utilization loci (PULs) from Bacteroides ovatus that targets the hemicellulose arabinoxylan, a heteropolymer with multiple monosaccharides and glycosidic linkages21. Unlike panel A, only the glycoside hydrolases in this locus are depicted, along with their predicted cellular locations: above the outer membrane (OM) are extracellular lipoproteins, between the OM and inner membrane (IM) are periplasmic enzymes and below the IM are cytoplasmic enzymes. A representation of maize arabinoxylan21is presented using the same monosaccharide scheme as presented in Fig. 1. The various B. ovatus glycan-degrading enzymes are color coded based on the linkages in maize arabinoxylan that they are predicted to degrade. Glycoside hydrolases are color-coded to represent the monosaccharide linkages hydrolyzed by each enzyme; labeling with two colors means that the enzyme family listed includes members capable of degrading two linkages present in arabinoxylan.