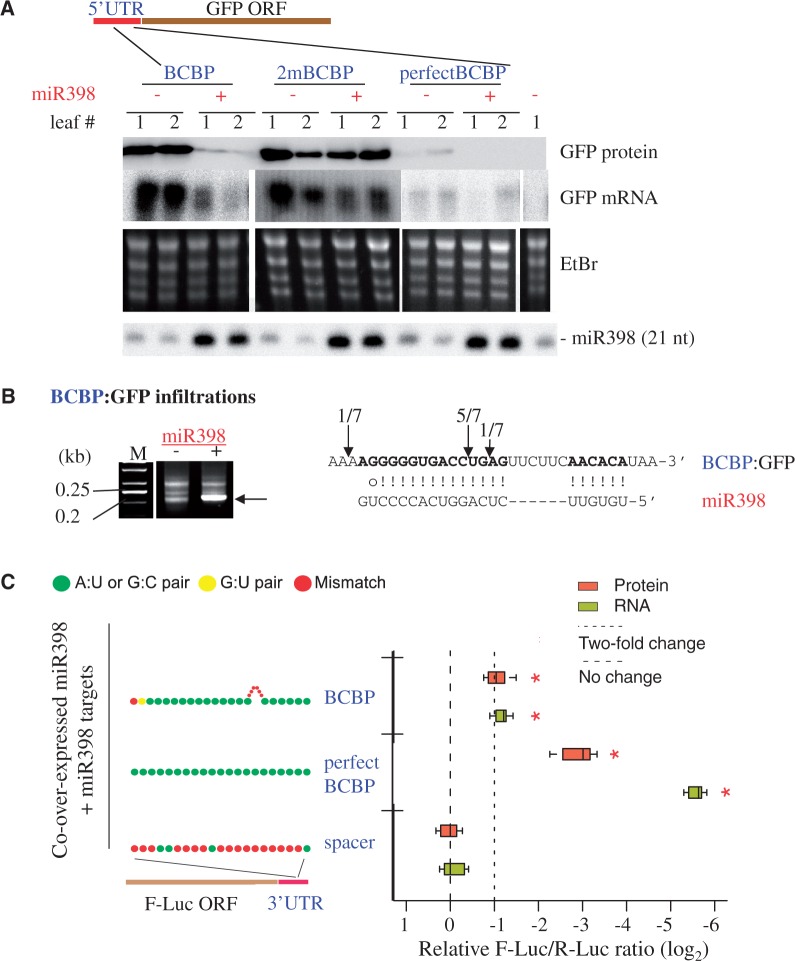
Figure 4.
The BCBP site functions outside of the BCBP mRNA context in both 5′- and 3′-UTRs. (A) BCBP 5′-UTR fusion with the GFP gene. Western and northern blot analyses to determine the levels of the GFP protein and mRNA in two independent N. benthamiana leaves (notated ‘1’ and ‘2’) infiltrated with the constructs indicated. The BCBP:GFP construct corresponds to the 5′-UTR of BCBP fused to the GFP. 2mBCBP has two mismatches around the cleavage site that disrupt the complementarity (Figure 3A). perfectBCBP is perfectly complementary to miR398. miR398 was detected with a radiolabeled oligonucleotide probe. (B) 5′-RACE PCR analyses of the BCBP site fused to the 5′-UTR of GFP. Total RNAs from N. benthamiana leaves infiltrated with the BCBP:GFP construct were analyzed by 5′-RACE PCR. The arrows indicate the DNA fragments cloned and sequenced to map the cleavage sites (on the right) located within the 5′-UTR. M; GeneRuler 50-bp DNA ladder (Fermentas). (C) BCBP 3′-UTR fusion with the F-Luc gene. Boxplots summarize results from nine independent replicates and show the median (thick line), extent of the 1st to 3rd quartile range (box), values extending to 1.5 times the interquartile range (whiskers), and outliers (black circles). Asterisks indicate significant differences comparing with the spacer control (P < 0.05, analysis of variance–Tukey’s honest significance difference). Colored dots show the schematic base-pairing pattern between miR398 and its targets with the target 5′ ends on the left.