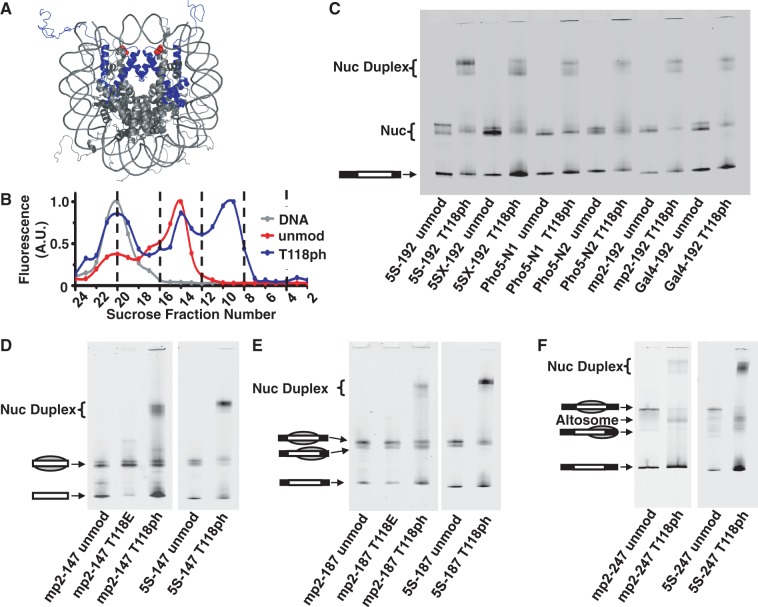
Figure 1.
Phosphorylated H3(T118) forms altered DNA–histone complexes. (A) Nucleosome crystal structure (26) with histone H3 in blue and H3(T118ph) in red. (B) Integrated fluorescence intensity of each fraction for sucrose gradient purification of mp2-247 DNA alone (gray), mp2-247 reconstituted with unmodified histone octamer (HO) (red) and mp2-247 reconstituted with HO containing H3(T118ph) (blue). Fraction numbers in (B) correspond to the fractions resolved by EMSA in Supplementary Figure S1C for the H3(T118ph) sample. (C) EMSA of NPS-192 DNAs reconstituted with either unmodified or H3(T118ph) histone octamer. (D) EMSA of DNA–histone complexes with NPS-147 DNAs containing either the mp2 or L. variegatus 5S NPS reconstituted with unmodified, H3(T118E) or H3(T118ph) HO. The gray sphere superimposed on the DNA pictogram indicates a nucleosome and its position on the DNA. (E) EMSA of NPS-187 DNA reconstituted with unmodified, H3(T118E) or H3(T118ph) HO. (F) EMSA of NPS-247 DNAs reconstituted with unmodified HO or HO containing H3(T118ph).