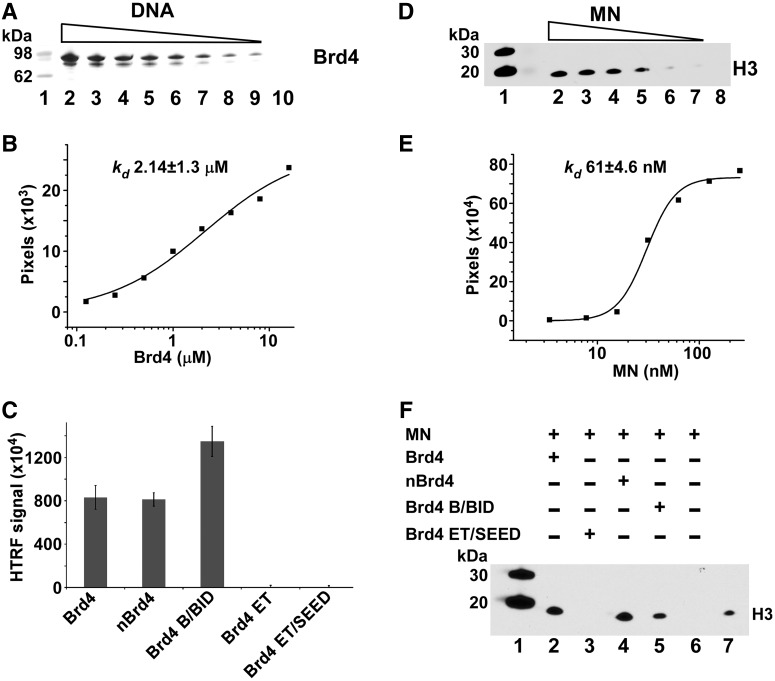
Figure 7.
Brd4 interactions with DNA (A, B and C) and MNs (D, E and F). (A) Streptavidin linked sepharose bead-based pull-downs of decreasing concentrations (lanes 2–9) of purified recombinant 6xHis–Brd4(1–720) with biotinylated 40-bp DNA were separated by SDS-PAGE and visualized by Coomassie Blue staining. The control to rule out non-specific binding is shown with 125 nM 6xHis–Brd4(1–720) minus biotinlyated DNA (lane 10). Molecular weight markers are in lane 1. (B) Experiments were run in triplicate with very similar results with a graphical representation of one gel shown to determine an apparent binding Kd. The intensities of 6xHis–Brd4(1–720) bound to biotinlyated DNA were quantified using ImageJ software and data were fit to the Hill equation. (C) Interactions of indicated Brd4 domains with the biotinylated DNA were measured by the HTRF-based assay. Reactions were run in triplicate with bars representing standard deviations. (D) Nickel bead-based pull-down of purified native MNs with recombinant 6xHis–Brd4(1–720) and immunoblotting with H3 antibodies. A gradient of decreasing MNs (lanes 2–7) was run. Control pull-down is shown with 125 nM MNs without 6xHis–Brd4(1–720) (lane 8) to rule out non-specific binding to nickel beads. Native MNs contain naturally occurring histone modifications and cellular genomic DNA. (E) Graphical representation of immunoblot is also shown to determine an apparent binding Kd. The intensities of H3 bands were quantified using ImageJ software and data were fit to the Hill equation. (F) Nickel bead-based pull-down reactions with indicated fragments of 6xHis–Brd4 and MNs. Bound MNs were detected by immunoblotting with anti-H3 antibody.