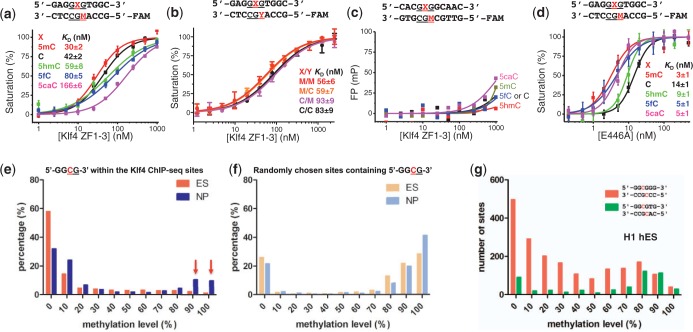
Figure 2.
The effects of CpG modifications and DNA sequence on DNA-binding by Klf4. (a) Binding affinities measured by fluorescence polarization assays between Klf4 and DNA with five different modification states on the upper strand (5mC = M, C, 5hmC, 5fC and 5caC). (b) Binding affinities measured between Klf4 and DNA that is fully methylated, unmodified or hemimethylated (on either strand). For these experiments, only M (5mC) and C were used. (c) A GCG-containing DNA sequence partially matching the consensus binding element of Klf4 (underlined) was used as a negative control. Fluorescence polarization (FP) is measured in millipolarization (mP). (d) Binding affinities measured between the E446A variant of Klf4 and DNA having five different modification states on the top strand. In all cases, the lower strand has M (5mC). (e) Distribution of DNA CpG methylation in mouse ES cells and NP cells (33) that is present within the core GGCG Klf4-ChIP sites identified in ES cells (23). The red arrows indicate changes of methylation levels, from hypomethylation in mouse ES cells to hypermethylation in NP cells (see Table 2), during differentiation and/or reprogramming. (f) Distribution of DNA methylation of randomly chosen GGCG sites in the mouse ES genome. (g) Distribution of DNA methylation in human H1 ES cells presented within the human KLF4-ChIP sites (34). GGCGTG sequences (green) have a higher proportion of methylated sites than do GGCGGG sequences.