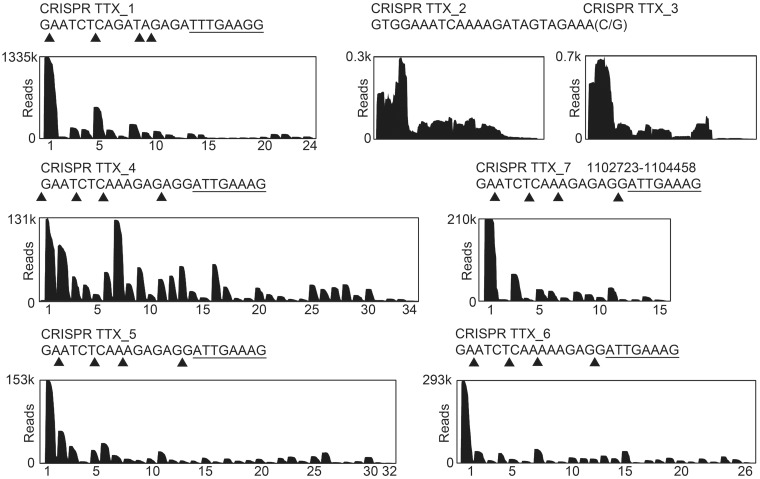
Figure 1.
Processing of crRNAs in T. tenax. The abundance of crRNAs was verified by mapping the Illumina HiSeq2000 sequencing reads to the T. tenax Kra1 reference genome. Sequence coverage (reads) for the seven T. tenax CRISPR clusters (crRNAs are indicated by numbers on the x-axis) is visualized. The processing sites could be identified within the repeat elements, generating crRNAs with a 5′-terminal 8-nt tag [5′-(A/U)UUGAA(A/G)G-3′, underlined] and variable trimming of the 3′-ends with most of the crRNAs terminating with a 1–2-nt tag (5′-G/GA-3′). The trimming sites for each CRISPR locus are indicated by triangles.