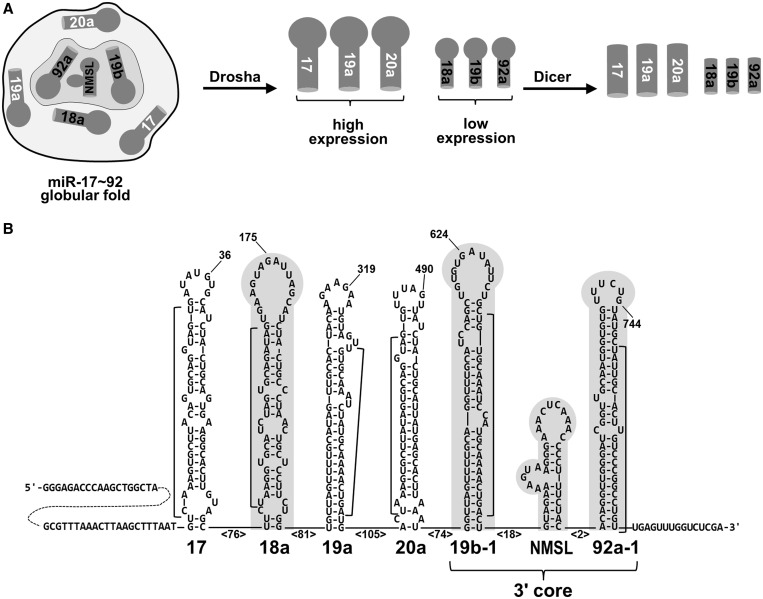
Figure 1.
Structural regulation of miR-17∼92 miRNA processing. (A) The globular fold of the miR-17∼92 pri-miRNA internalizes miR-19b and miR-92a of the 3′ core domain as well as miR-18a. The internalized miRNAs have reduced expression relative to surface exposed miR-17, miR19a and miR20a due to suppressed Drosha processing. (B) Predicted secondary structure (mFold) of the miR17∼92 cluster. The sequence shown is that of the T7 RNA polymerase run-off transcript generated from Xho I–digested pcDNA 3.1 (+) vector. Select nucleotides are numbered for reference, the numbers in angle brackets indicate the number of intervening nucleotides. The internalized 3′ core domain and miR-18a hairpin are shaded gray and mature miRNA sequences are indicated with a bracket. Nucleotides 678–710 comprise the non-miRNA containing stem-loop (NMSL).