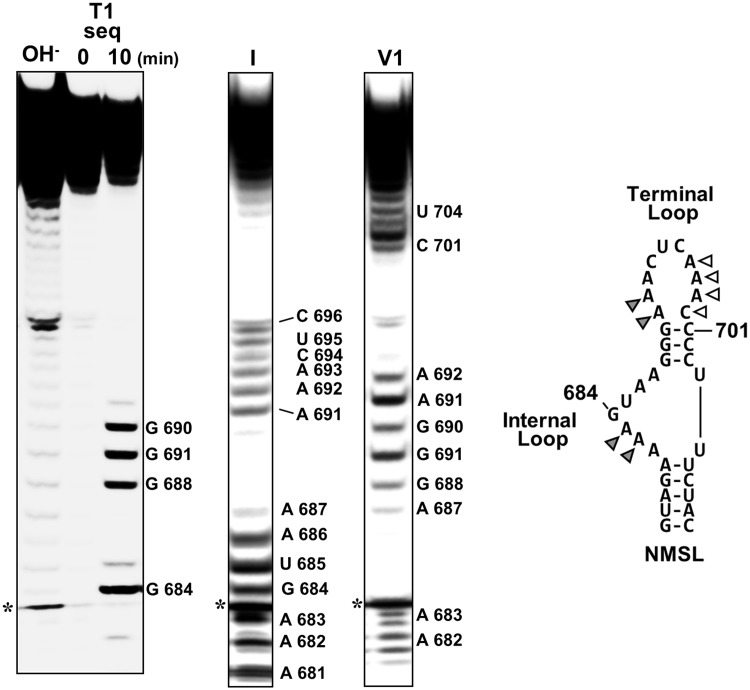
Figure 3.
Structure probing of the NMSL. Denaturing PAGE analysis of RNAse 1 and RNAse V1 cleavage of 5′-[32P]-end-labeled NMSL. Cleavages were assigned using a base hydrolysis ladder and an RNAse T1 sequencing reaction. Adenosines that are cleaved by both RNase 1 and V1 are indicated by gray triangles. The white triangles indicate positions that are not cleaved by either RNase. The predicted Watson–Crick base pairs A681:U705 and A687:U704 could not be confirmed by ribonuclease sensitivity. The asterisk indicates a nonribonuclease degradation product. 3′ heterogeneity of the low molecular weight cleavage products for positions A 682 and A 683 are visible.