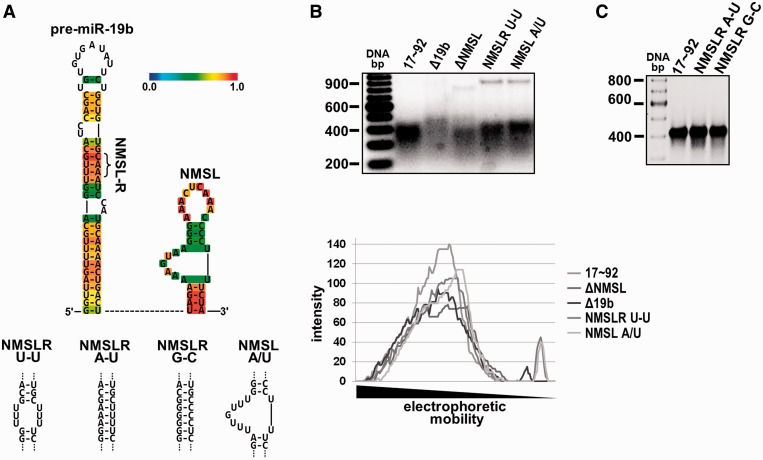
Figure 5.
Sequence dependence of the NMSL-NMSLR interaction. (A) Top panel, predicted miR-19b secondary structure using CentroidHomfold (39). The color scale bar indicates base-pairing probability. Base-pairs in the stem have the highest probability. Bottom panel, miR-19b stem and NMSL mutants used in (B) and (C). (B) Top panel, deletion of the NMSL or miR-19b stem, or mutation of the adenosine repeats of the NMSL internal loop or the NMSLR yield nonhomogenous populations of folded RNAs. Bottom panel, quantification (ImageQuant) of band intensity of the native gel lanes (C) Restoring the A/U base pairing of the NMSLRU-U mutant with A/U or G/C base pairing results in homogenous populations of folded RNAs. 2.5 mM MgCl2 in the gel and running buffer, 5 µg RNA was loaded in each lane in both (B) and (C).