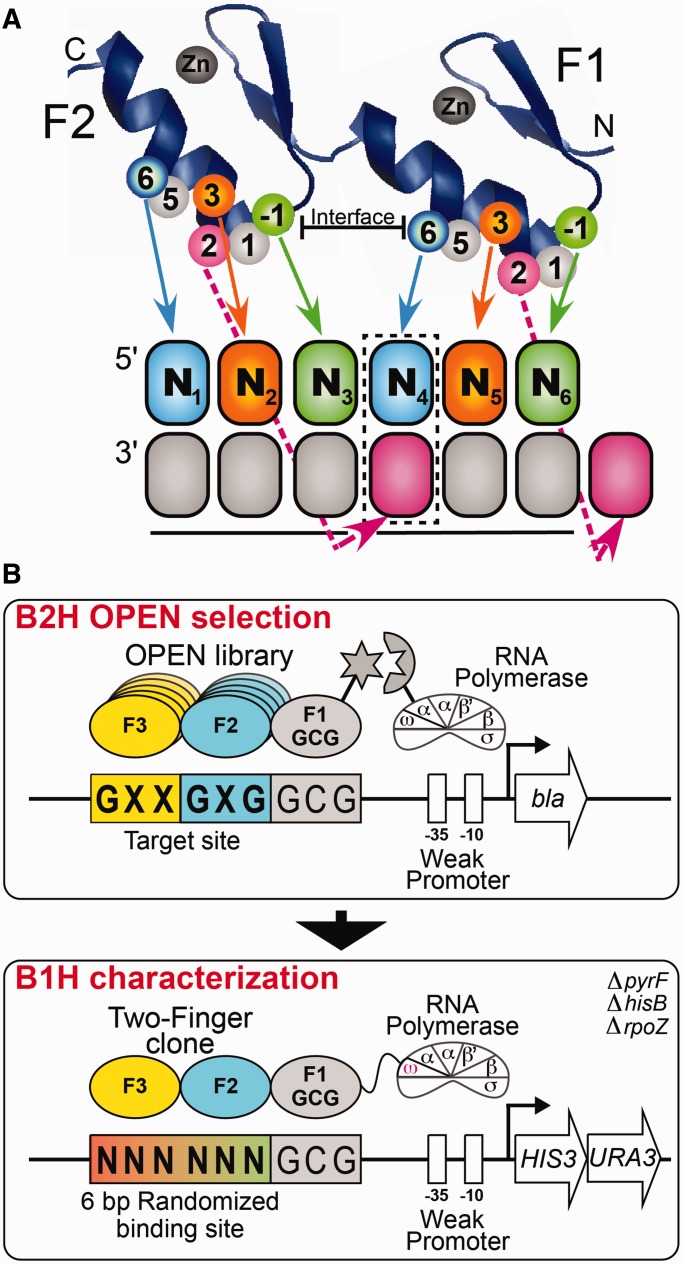
Figure 1.
(A) Schematic representation of the canonical recognition pattern of two zinc fingers recognizing a hexamer sequence. Each zinc finger unit spans ∼30 amino acids and folds into a ββα-motif around a tetrahedrally coordinated zinc ion (42,43). DNA-binding specificity is typically mediated by residues at positions −1, +2, +3 and +6 of the recognition helix, where the numbering scheme refers to the position of each residue relative to the start of the α-helix. The boxed base pair (N4) represents the position of potential recognition overlap in the canonical recognition model. (B) Schematic representation of the two-stage process used to identify two-finger modules with the desired sequence preference. In Stage 1, the B2H system is used to select two-finger modules from an OPEN-based library, where the finger pools used correspond to the finger 2 (F2) and finger 3 (F3) subsites in each target site (44,45). These two-finger libraries are selected in the context of a constant finger 1 (F1) module that recognizes GCG in the neighboring subsite. The DNA-binding specificity of active clones recovered from the B2H selection was determined using the B1H system using a 6-bp randomized library adjacent to the constant GCG F1 binding site. The recovered binding sites are determined by Illumina sequencing and then a binding site motif is calculated from these sequences (46).