Abstract
Background
Cysteine, a sulfur-containing amino acid, plays an important role in a variety of cellular functions such as protein biosynthesis, methylation, and polyamine and glutathione syntheses. In trypanosomatids, glutathione is conjugated with spermidine to form the specific antioxidant thiol trypanothione (T[SH]2) that plays a central role in maintaining intracellular redox homeostasis and providing defence against oxidative stress.
Methods
We cloned and characterised genes coding for a cystathionine β-synthase (CβS) and cysteine synthase (CS), key enzymes of the transsulfuration and assimilatory pathways, respectively, from the hemoflagellate protozoan parasite Trypanosoma rangeli.
Results
Our results show that T. rangeli CβS (TrCβS), similar to its homologs in T. cruzi, contains the catalytic domain essential for enzymatic activity. Unlike the enzymes in bacteria, plants, and other parasites, T. rangeli CS lacks two of the four lysine residues (Lys26 and Lys184) required for activity. Enzymatic studies using T. rangeli extracts confirmed the absence of CS activity but confirmed the expression of an active CβS. Moreover, CβS biochemical assays revealed that the T. rangeli CβS enzyme also has serine sulfhydrylase activity.
Conclusion
These findings demonstrate that the RTS pathway is active in T. rangeli, suggesting that this may be the only pathway for cysteine biosynthesis in this parasite. In this sense, the RTS pathway appears to have an important functional role during the insect stage of the life cycle of this protozoan parasite.
Keywords: Cysteine biosynthesis, Cystathionine β-synthase, Cysteine synthase, T. rangeli, Thiol metabolism, Antioxidant defence
Background
L-cysteine, a sulfur-containing amino acid, is indispensable for the survival of virtually all living organisms, from bacteria to higher eukaryotes. This amino acid is implicated in several processes, including the stability, structure, regulation of catalytic activity, and post-translational modification of various proteins [1]. Due to the ability of its thiol group to undergo redox reactions, L-cysteine forms the basic building block of all thiol antioxidants, acting as a direct antioxidant and also as a precursor for the biosynthesis of glutathione, trypanothione, or ovothiol [2]. In addition, cysteine is also essential for the synthesis of biomolecules, including coenzyme A, hypotaurine, taurine, and ubiquitous iron-sulphur (Fe-S) clusters, which are involved in electron transfer, redox regulation, nitrogen fixation, and regulatory process sensing [3,4].
Two different routes for cysteine biosynthesis have been described: reverse-transsulfuration (RTS) and de novo or assimilatory pathways. RTS has been demonstrated in fungi and mammals and includes the complete process leading to cysteine from methionine via the intermediary formation of cystathionine [5]. These reactions are catalysed by two enzymes, CβS (cystathionine β-synthase), which synthesizes cystathionine from homocysteine and serine, and CGL (cystathionine γ-lyase), which forms cysteine from cystathionine [6]. The de novo pathway is also catalysed by two steps starting with serine acetyltransferase (SAT) to form O-acetylserine (OAS) from L-serine and acetyl-coenzyme A. Subsequently, OAS reacts with sulfide to produce cysteine in an alanyl-transfer reaction by cysteine synthase (CS) [7]. This de novo pathway for cysteine biosynthesis is found in plants, bacteria, and some protozoa, such as Entamoeba histolytica, Entamoeba dispar [8], Leishmania major [9], and Leishmania donovani [10], but is absent in mammals [11]. Both CβS and CS are PLP-dependent enzymes that are evolutionary-related and in most cases some CS activity has been demonstrated for the CβS enzymes described to date [12].
It is well established that the antioxidant defence system plays a key role in the host-parasite interaction for intracellular pathogenic trypanosomatids such as T. cruzi and Leishmania spp., promoting the protection of the parasite against macrophage-derived oxygen and nitrogen-reactive species [13,14]. Among trypanosomatids, the mammalian-infective and non-pathogenic Trypanosoma rangeli is of growing interest because its intracellular life stage within mammalian hosts is still unknown and its sympatric occurrence with T. cruzi [15].
Because T. rangeli is required for a response to a variety of oxidative stresses in both mammalian and invertebrate hosts, the present study characterised genes encoding key enzymes of cysteine biosynthesis, a crucial precursor of trypanothione.
Methods
Parasites and culture
Epimastigotes of T. rangeli Choachí strain and T. cruzi Y strain were grown at 26.5°C in liver infusion tryptose medium (LIT) supplemented with 10% heat-inactivated fetal bovine serum (FBS), 100 units/mL penicillin, and 100 μg/mL streptomycin by weekly passaging [16]. Parasites were harvested at the late log phase for DNA or protein extraction as well as for thiol profiling and in vitro oxidative and nitrosative stress testing. Trypomastigotes of T. rangeli were obtained in vitro under conditions previously described [17].
T. cruzi culture-derived trypomastigotes and amastigotes were obtained from THP-1 differentiated macrophage-like cells (ATCC) infected with Y strain metacyclic trypomastigotes [18]. Briefly, THP-1 cells (ATCC) were cultured in RPMI 1640 medium supplemented with 10% FBS at 37°C in a 5% CO2 atmosphere and transformed to adherent macrophages using phorbol myristate acetate (50 ng/mL) for 72 h at 37°C and 5% CO2 prior to experiments. THP-1 macrophage-like cells were infected with T. cruzi trypomastigotes for 2 h at a 3:1 parasite-cell ratio and then washed to remove the extracellular parasites. After 72 h at 37°C under 5% CO2, the trypomastigotes were collected from the culture supernatant, centrifuged at 600 × g for 30 min, and then left under the same conditions for 3 h to separate the trypomastigotes from the amastigotes and cellular debris. The supernatant containing the trypomastigotes was used for protein extraction.
Identification of T. rangeli CβS and CS
Both the T. rangeli genome and transcriptome databases (http://www.rangeli.lncc.br) [19] were searched using the TBLASTN algorithm with the protein sequences of cystathionine β-synthase (CβS) and cysteine synthase (CS) from bacteria, yeast, plants, and parasitic protozoa as queries to identify putative T. rangeli proteins involved in transsulfuration and assimilatory pathways. Other coding sequences for potential enzymes comprising the two biosynthetic pathways were also searched in the genome and transcriptome databases. T. rangeli genomic DNA (gDNA) was isolated by the phenol–chloroform method following a standard protocol [20]. The open reading frames (ORFs) of the CβS and CS genes were amplified by PCR using gene-specific primers: CBTrXhoI (5′-CTC GAG ACC ATG GCT CAA ACC CAC-3′) and CBTrBamHI (5′-GGA TCC GCG CAC CTG CTT TTT ATC C-3′) for CβS and CSTrNdeI (5′- CAT ATG GAA GCT CTC ATC GGG G-3′) and CSTrXhoI (5′- CTC GAG CCA GCA CCA CGG GAA GC-3′) for CS. Sites for restriction enzymes (included in the primer name; bolded nucleotides) were included to allow cloning. All PCR assays were carried out using a Mastercycler® Gradient (Eppendorf, Hamburg) for 30 cycles of denaturation (94°C, 1 min), annealing (60°C, 45 sec), and extension (72°C, 1 min), followed by a final extension step (72°C) for 5 min. The PCR products were cloned into the pGEM-T-Easy vector (Promega), and the resulting constructs were verified by sequencing using a Megabace 1000® DNA Analysis System with the DYEnamic ET terminators kit (GE Healthcare) according to the manufacturer’s conditions. Both DNA strands were sequenced for each clone obtained; after analysis using the Phred/Phrap/Consed package [21], only high-quality DNA sequences (Phred ≥ 20) were compared with the public databases using the GenBank BLAST algorithm.
Protein expression and purification
The inserts corresponding to the CβS and CS ORFs cloned into pGEM-T-Easy (Promega) were excised and subcloned into the pET14b expression vector (Novagen) pre-digested with the appropriate restriction enzymes (included in the PCR primers). The resulting plasmids containing the CβS and CS genes were named pET14-TrCβS and pET14-TrCS, respectively, and re-sequenced for confirmation as described above.
The pET14-TrCβS plasmid was used to transform E. coli BL21 (DE3) for recombinant protein expression. Pre-inoculum was grown overnight in LB (Luria– Bertani) broth supplemented with 100 μg/mL ampicillin at 37°C and then used to inoculate fresh LB until an O.D.600 of 0.6 was reached. The expression of recombinant CβS (rTrCβS) was induced with 1 mM isopropyl β-D-thiogalactopyranoside (IPTG) for 2 h at 37°C. The cells were harvested and resuspended in 5 mL of buffer A [50 mM sodium phosphate, 0.3 M NaCl, pH 8.0, and 25 μM pyridoxal phosphate (PLP)] containing 5 mM imidazole and then disrupted by sonication. The soluble and insoluble fractions were recovered by centrifugation at 16,000 × g for 30 min at 4°C [9]. rTrCβS was purified from insoluble fractions by affinity chromatography on a Ni2+-nitrilotriacetic (NTA) column (Qiagen) following standard procedures. Briefly, the insoluble fraction was resuspended in a buffer containing 8 M urea, 10 mM Tris, and 100 mM NaH2PO4, pH 8.0, and incubated for 1 h at 65°C to dissolve the inclusion bodies and then centrifuged (10,000 × g for 30 min at 4°C). The supernatants were then applied to the Ni2+-nitrilotriacetic (NTA) resin (Qiagen) pre-equilibrated with the same buffer and incubated for 1 h at 4°C under continuous agitation. The resin was washed three times using washing buffer (100 mM NaH2PO4, 100 mM Tris/HCl, and 8 M urea, pH 6.3), and rTrCβS elution was carried out using an appropriate buffer (100 mM NaH2PO4, 100 mM Tris/HCl, and 8 M urea, pH 4.5). The eluted proteins were dialysed using 50 mM NaH2PO4 pH 7.4, 300 mM NaCl and 150 mM imidazole overnight at 4°C. The purity of the recombinant protein was then assessed by SDS-PAGE, and its concentration was determined by the Bradford method (Bio-Rad) using BSA as a standard. The protein was stored at -20°C.
To obtain recombinant CS (rTrCS), different approaches were assessed. pET14-TrCS was introduced into E. coli BL21 (DE3), BL21 (DE3)pLysS, and Rosetta strain, and expression was induced using different IPTG concentrations (0.2, 0.5, or 1.0 mM) and temperatures (15°C, 25°C, or 37°C). Despite the number of experimental conditions tested, it was not possible to obtain recombinant TrCS.
Production of α-rTrCβS mouse polyclonal antibodies
Approximately 50 μg of purified rTrCβS (44 kDa) was subcutaneously inoculated into Balb/C mice using Alu-Gel (Serva) as an adjuvant. Each mouse received four consecutive inoculations at 12-day intervals, with monitoring for an antibody response using enzyme-linked immunosorbent assay (ELISA) with rTrCβS as the antigen.
Comparative analysis of CβS expression by T. rangeli and T. cruzi
Quantification of CβS expression was performed using soluble protein fractions from T. rangeli and T. cruzi. A total of 1 × 108 epimastigotes or trypomastigotes were washed once with D-PBS and lysed by repeated aspiration in ice-cold lysis buffer (0.25 M sucrose, 0.25% Triton X-100, and 10 mM EDTA) containing a protease inhibitor cocktail (Sigma-Aldrich). Cellular debris was removed by centrifugation at 12,000 × g for 20 min at 4°C [22]. The protein concentrations in the extract were determined by the Bradford method (Bio-Rad) using BSA as a standard and stored at -20°C.
Soluble protein extracts (30 μg) of the different life cycle stages of T. rangeli and T. cruzi were fractionated on 12% SDS-PAGE and electroblotted onto nitrocellulose membranes (GE Healthcare) in an appropriate buffer (25 mM Tris; 192 mM glycine; 20% v/v methanol, pH 8.3). The membranes were then blocked with 5% non-fat milk in blotting buffer (25 mM Tris–HCl pH 7.4, 150 mM NaCl, and 0.1% Tween-20) overnight at 4°C [23]. After blocking, the membranes were incubated for 1 h with an anti-rTrCβS mouse polyclonal antiserum (1:4,000) or anti-α tubulin monoclonal antibody (1:10,000) used as a loading control. After washing, the membranes were incubated with anti-mouse IgG conjugated to horseradish peroxidase (1:10,000), followed by washing and detection on radiographic films using an ECL kit (Pierce) according to the manufacturer’s recommendations. The western blots were digitally analysed using the software package Image J 1.463r, subtracting the background of each blot prior to measuring the intensity of specific bands. Integrated densities for each band were determined for each protein of interest and its corresponding loading control. The ratio of the band intensity of the protein of interest versus the band intensity of the corresponding loading control was used as the relative protein expression level and allowed the comparison with other samples.
Enzymatic assays for CβS and CS activities
Cystathionine β-synthase
The assay method described by Walker and Barret was used [24]. Briefly, the reaction mixture contained 70 μmol Tris–HCl buffer (pH 8.4), 0.4 mM PLP, and 1.5 μg/μL of total protein extract from parasites or 0.1 μg/μL of rTrCβS (as a positive control) in a final volume of 100 μL. In the case of the protein extract, the mixture also contained 0.1 mM CuSO4 to inhibit cystathionase activity. All components were equilibrated for 2 min at 37°C, and the reaction was initiated by the addition of 40 mM D,L-homocysteine and 20 mM L-serine. The reaction was stopped 45 min later by the addition of 100 μL 50% (w/v) trichloroacetic acid. The precipitated protein was removed by centrifugation at 12,000 × g for 5 min, and the amount of cystathionine was determined by adding 1 mL of acid-ninhydrin reagent (1 g ninhydrin dissolved in 100 mL concentrated acetic acid and 1/3 volume of phosphoric acid) to 100 μL of the assay supernatant fraction. The mixture was then boiled for 5 min, cooled for 2 min on ice, and incubated for 20 min at room temperature (25°C) for colour development. The absorbance was measured at 455 nm. Each enzymatic assay was performed including negative controls (all reagent components without enzyme or without substrate). A standard curve was prepared using 0–3 μmol of cystathionine dissolved in acid-ninhydrin reagent and treated as described above to quantify the amount of cystathionine formed [25].
Cysteine synthase
The CS activity in the total protein extracts from parasites (1.5 μg/μL) or bacteria (positive control) was determined by measuring cysteine production at 37°C in a 500 μL reaction containing 200 mM potassium phosphate buffer (pH 7.5), 10 mM DTT, 0.2 mM PLP, 6.5 mM O-acetylserine (OAS), and 4 mM sodium sulfide (Na2S). All the components except sodium sulfide were pre-incubated for 5 min at 37°C; the reaction was initiated by the addition of sodium sulfide and incubated for another 30 min and then stopped using 50 μL of 20% trichloroacetic acid (w/v). The mixture was centrifuged for 5 min at 12,000 × g, and the supernatant was used for cysteine analysis, as previously described with some modifications [26]. Briefly, an aliquot (500 μL) of the supernatant was added to 500 μL of ninhydrin reagent (250 mg ninhydrin dissolved in 10 mL concentrated acetic acid: concentrated HCl, 60 ~ 40 v/v). The mixture was boiled for 10 min and immediately cooled on ice before the addition of 500 μL of 95% (v/v) ethanol. The amount of cysteine formed was determined by measuring the absorbance of the reaction mixture at 560 nm [27]. Each enzymatic assay was performed including negative controls (all reagent components without enzyme or without substrate). A standard curve was prepared with L-cysteine (0–1 μmol) dissolved in ninhydrin reagent and treated as described above to quantify the amount of cysteine formed. The serine sulfhydrylase activity of CS was determined in the same way as described for the CS assay above, except that 6.5 mM serine was used instead of OAS.
Cellular thiol contents
The total thiol content of T. rangeli and T. cruzi epimastigotes was determined using deproteinised parasite extracts prepared as formerly described [28]. Epimastigotes in the exponential phase (1 × 108 parasites/mL) were harvested, washed with D-PBS, and suspended in 0.6 mL of 25% trichloroacetic acid. After 10 min on ice, the denatured proteins and cell debris were removed by centrifugation at 13,000 × g for 10 min at 4°C. The thiol content of the supernatant solution was determined by Ellman’s method [29] using 0.6 mM 5,5′-dithio-bis(2-nitrobenzoic acid) (DTNB) in 0.2 M sodium phosphate buffer (pH 8.0). The concentration of DTNB derivatives of thiols was estimated spectrophotometrically at 412 nm. Calibration curves were performed with known amounts of cysteine.
Epimastigote susceptibility to oxidative and nitrosative stress in vitro
Parasite susceptibility to oxidative or nitrosative stress was assessed using Alamar blue (AB) assays, as described elsewhere [22,30] with minor modifications. Briefly, 5 × 105 T. rangeli and T. cruzi epimastigotes were incubated for 48 h with 100 μL parasite culture in quadruplicate in 96-well plates. Aliquots of 100 μL of 30% hydrogen peroxide (Sigma-Aldrich) or S-nitroso-N-acetylpenicillamine (SNAP, Molecular Probes®- Life Technologies) prepared at different dilutions (0–100–150–300–500–1000–1500 μM and 0–5–20–50–150–300–500–1000 μM, respectively) were added, as reported [22,31]. After incubation at 26°C for 24 h, 20 μL of AB reagent (Invitrogen) was added to each well to assess parasite viability via fluorescence emission at 600 nm. Data from treated and non-treated cultures were used to calculate the IC50 by a sigmoidal regression analysis (with variable slope) using GraphPad Prism v.5.0. Untreated control parasites and reagent blanks were included in each test plate.
Statistical analysis
All experiments were performed in triplicate, and the results are presented as the mean and the standard deviation (SD) or standard error of the mean (SEM). Normalised data were analysed by a one-way ANOVA followed by Bonferroni post-tests or Student’s t-test, as indicated in the figure legends, using the software GraphPad Prism v.5.0.
Ethical approval
The procedures involving animals were previously approved by the UFSC Ethics Committee on Animal Use – CEUA (Reference number: 23080.025618/2009-81).
Results
The T. rangeli genome contains genes encoding CβS and CS enzymes
Using the nucleotide and protein sequences of CβS and CS orthologs from plants, bacteria, yeast, and parasitic protozoa as queries, a search of T. rangeli genome and transcriptome databases allowed the identification of genes encoding CβS and a partial gene sequence for CS. Additionally, the T. rangeli genome contains a single copy of the cystathionine γ-lyase (CGL) gene of the RTS pathway but lacks the genes encoding serine acetyltransferase (SAT) present in the de novo biosynthetic pathway of other trypanosomatids. The sequences for CβS and CS were then back-searched using the SWISSPROT and NCBI databases, which confirmed the identity of both genes. These results suggest that, as in other trypanosomatids, T. rangeli possesses genes coding for the enzymes involved in these two cysteine biosynthetic routes: CβS in the transsulfuration pathway and CS in the de novo biosynthesis pathway.
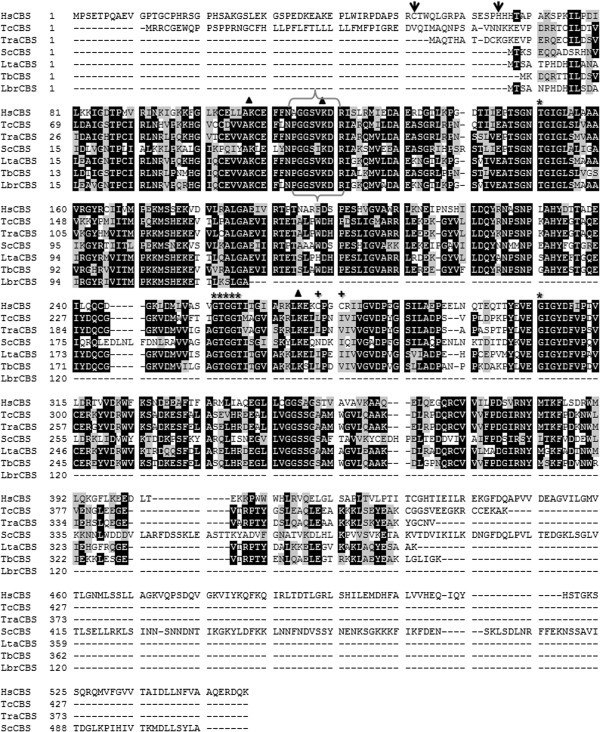
After cloning and sequencing, it was found that T. rangeli CβS (TrCβS) predicts a protein of 373 amino acids (44 kDa) that reveals high sequence identity with CβS from T. cruzi (84%), T. brucei (78%), and L. major (75%) compared to human CβS (50%). Multiple sequence alignment confirmed that TrCβS contains three out of the four lysine residues (Lys 53, Lys64, Lys213) required for CS activity; the consensus sequence for the putative cofactor pyridoxal phosphate-binding domain is highly conserved. rTrCβS, as well as CβS from other trypanosomatids, differs from H. sapiens CβS (HsCβS) by lacking the haem-binding (redox sensor) and oxidoreductase motifs (Cys XX Cys) at the N- and C-termini, respectively (Figure 1).
Figure 1.
Multiple alignment of deduced amino acid sequences of CβS from T. rangeli (TrCβS) and other representative organisms. The identity (black background) and conservation (grey background) of the amino acid residues are shown. Brackets indicate the consensus amino acid residues of the putative pyridoxal phosphate-binding motif (PXXSVKDR), and other motifs vital for CβS activity are indicated with asterisks (*). The oxido-reductase motif of HsCβS is highlighted with (+). The lysine residues required for CS catalytic activity are marked with triangles. The positions of the heme-binding residues within the heme domain of the human CβS enzyme (Cys52 and His65) are marked with (↓). HsCβS: Human (P35520); TcCβS: Trypanosoma cruzi (Tc00.1047053511691.20); ScCβS: Saccharomyces cerevisiae (P32582) LtaCβS: Leishmania tarentolae (LtaP17.0270); TbCβS: Trypanosoma brucei (Tb11.02.5400); LbrCβS: Leishmania braziliensis (LbrM.17.0230).
The T. rangeli CS gene (TrCS) encodes a protein of 155 amino acids (~16.8 kDa) that is 53% identical to the T. cruzi ortholog but exhibits lower identity with L. major (46%) and L. infantum (45%). Although CβS and CS are evolutionarily related enzymes, we found a low identity between TrCβS and TrCS (≤13%) when compared to the TrCS identity with the other orthologues from plants and bacteria (~31-33%). An analysis of the predicted amino acid sequences of TrCS revealed an amino acid change of Pro32 → Ser within the putative pyridoxal phosphate-binding domain (PXXSVKDR). Unlike other CSs, TrCS has only two of the four lysine residues (Lys37, Lys 53) shown to be important for the catalytic activity of the enzyme. Furthermore, TrCS does not have the canonical β8-β9 loop described in CS enzymes, which is important for access to the active site, and neither of the positively charged residues (Lys-His-Lys) involved in binding with serine acetyl-transferase (SAT) (Figure 2).
Figure 2.
Multiple alignment of deduced amino acid sequences of CS from T. rangeli and other representative organisms. The identity (black background) and conservation (grey background) of the amino acid residues are shown. Brackets indicate the consensus amino acid residues of the putative pyridoxal phosphate-binding motif (PXXSVKDR); the substitute for the proline residue is marked with (•), and the lysine residues required for cysteine synthase activity are indicated with triangles. The β8–β9 loop at the entrance of the active site is indicated with an asterisk (*), and the positively charged residues involved in binding with SAT are indicated with (+). TraCS: Trypanosoma rangeli; TcCS: Trypanosoma cruzi (Tc00.1047053507165.50); LbrCS: Leishmania braziliensis (LbrM.35.3820); LmjCS: Leishmania major (LmjF.36.3590); EhCS: Entamoeba histolytica; TvCS: Trichomonas vaginalis (XP001325874); StCS A: Salmonella typhimurium CysK (P0A1E4); StCS B: Salmonella typhimurium CysM (NP_456975).
Stage-specific expression of CβS in T. rangeli
The relative abundance of the CβS protein was evaluated in T. rangeli epimastigote and trypomastigote forms by western blotting, showing no significant differences between the forms. The absence of TrCβS stage-specific expression contrasts with the homologous protein in T. cruzi (TcCβS), for which the expression level of CβS was found to be significantly increased in epimastigotes (Figure 3A, B).
Figure 3.
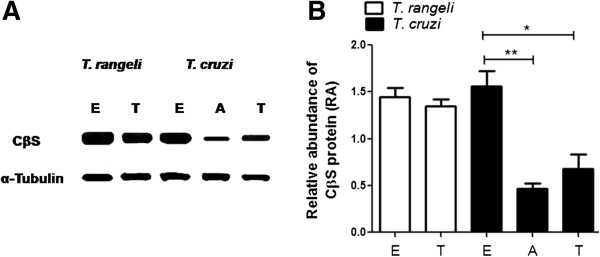
Protein expression levels of CβS in T. rangeli and T. cruzi. A. Western blot analysis of soluble extracts obtained from epimastigotes (E), and trypomastigotes (T) of T. rangeli and T. cruzi, and amastigotes (A) of T. cruzi. B. Densitometric analysis of CβS expression using ImageJ software and significant differences in CβS expression between epimastigotes and trypomastigotes, as determined by the t-test (*P < 0.05, **P < 0.01). The normalisation of protein loading was performed by the immunodetection of α-tubulin.
CβS is active in T. rangeli
The enzymatic studies on T. rangeli extracts showed that CβS activity is detectable in both epimastigotes (0.13 μmol min-1 mg-1) and trypomastigotes (0.079 μmol min-1 mg-1 of protein) (Figure 4A), whereas CβS activity was 1.9 times higher in the extracts from T. cruzi epimastigotes versus trypomastigotes. Conversely, CS activity was undetectable in the protein extracts from both T. rangeli forms (Figure 4B).
Figure 4.
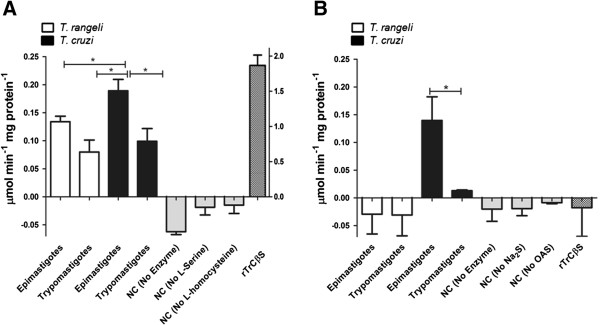
Detection of CβS and CS activities in protein extracts of T. rangeli and T. cruzi epimastigotes and trypomastigotes. A. The activities of CβS were determined in soluble extracts from trypanosomes using the recombinant enzyme rTrCβS as a positive control (axis Z). The results represent the average of five independent experiments performed in triplicate ± SD. B. The activities of CS were determined in soluble extracts from trypanosomes. The data represent the mean of five independent experiments performed in triplicate ± SD. Significant difference (*P < 0.05). NC = negative controls.
rTrCβS showed CβS activity of 2.2 ± 0.2 μmol min-1 mg-1 of protein (Figure 4A), with a km of 1.702 ± 0.11 mM for L-serine and a Km of 7.301 ± 1.9 mM for L-homocysteine, indicating a high binding affinity for L-serine and a weak binding affinity for L-homocysteine. rTrCβS was also capable of generating L-cysteine from serine and sodium sulfide, but with a very low specific activity (serine sulfhydrylase activity of 0.013 μmol min-1 mg-1 of protein). Different from T. cruzi CβS, rTrCβS did not show any CS activity (data not shown).
Total thiol content in T. rangeli and in vitro oxidative/nitrosative stress phenotyping
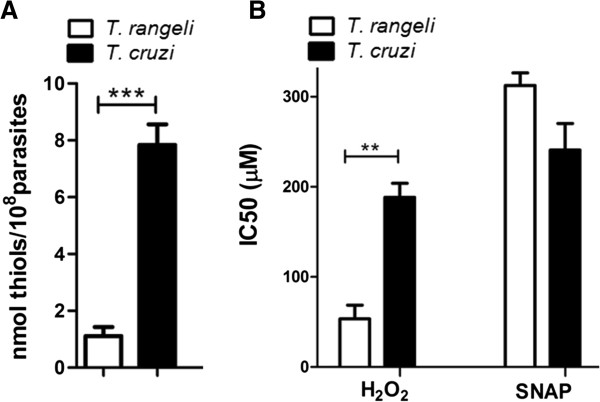
A comparative analysis of the total thiol levels of T. rangeli and T. cruzi revealed significant differences between these parasites. T. cruzi showed a thiol content of 7.8 nmoles/108 parasites, whereas T. rangeli had a thiol content that was almost seven times less (1.1 nmoles/108 parasites) (Figure 5A).
Figure 5.
Total thiol content and effects of oxidative and nitrosative stress on T. rangeli and T. cruzi viability. A. The total thiol content was determined in soluble extracts obtained from the epimastigote form. Error bars represent the SEM of three independent experiments. B. In vitro susceptibility of epimastigotes of T. rangeli and T. cruzi exposed to oxidative stress by hydrogen peroxide (H202) or nitrosative stress by S-nitroso-N-acetylpenicillamine (SNAP). Error bars represent the SEM of three independent experiments, performed in quadruplicate. Significant differences were detected by a one-way ANOVA, followed by Bonferroni post-tests (**P < 0.01, ***P < 0.001).
Based on these results, the T. rangeli susceptibility to oxidative and nitrosative stress was evaluated by subjecting epimastigotes to stress conditions in vitro with H2O2 or SNAP. This parasite was found to be more sensitive than T. cruzi to oxidative stress (H2O2), showing an IC50 of 53 μM, which is significantly less (P < 0.01) than the IC50 obtained for T. cruzi epimastigotes (188.3 μM). Nevertheless, the difference between these parasites was less pronounced under nitrosative stress conditions (SNAP), with T. rangeli being more resistant than T. cruzi (IC50: 312 μM and 240.7 μM, respectively) (Figure 5B).
Discussion
Our results indicate that RTS appears to be the only pathway for cysteine biosynthesis in T. rangeli. At the genomic level, T. rangeli contains single copies of genes coding for the CβS and CGL (cystathionine γ-lyase) enzymes of the RTS pathway but lacks genes encoding a protein of the cysteine de novo biosynthetic pathway (SAT). Additionally, a partial gene sequence for CS was found that has an A-G nucleotide transition at position 470, which generates a stop codon (TAG) (data not shown); thus, the truncated protein encoded lacks two of the four lysine residues required for CS activity.
A biochemical analysis of rTrCβS showed a higher CβS activity compared to hsCβS for generating cystathionine via the condensation of L-serine and L-homocysteine, though rTrCβS is less active than TcCβS [32-34]. In spite of this, the binding substrate affinity was comparable to the affinity of the CβS enzyme from L. major and humans [9]. Similar to other CβSs, rTrCβS can also form cysteine from L-serine and sodium sulfide, but is unable to utilise OAS and sulfide to catalyse the production of cysteine. Nevertheless, inter-species variations in other CβS catalysed reactions [24] could explain the absence of CS activity mediated by TrCβS.
The presence of a truncated CS gene as revealed by high-quality sequencing (Phred ≥50), and the absence of CS activity in both epimastigote and trypomastigote extracts suggests that the de novo cysteine biosynthetic pathway is absent or not functional in T. rangeli. Nevertheless, T. rangeli possesses a functional RTS pathway, a characteristic shared with T. brucei, for which only CβS activity has been reported in bloodstream trypomastigote extracts but at a very low level [35]. This result indicates that similarities in the metabolism of sulfur-containing amino acids exist between T. rangeli and T. brucei, another parasite that does not possess an intracellular mammalian host stage. Such findings may suggest that the extracellular stage of the life cycle of parasitic protozoa and the RTS biosynthetic pathway are causally connected.
No stage-specific association was found for T. rangeli CβS activity and protein levels, contrasting with T. cruzi, with epimastigotes (insect-form) that present significantly higher activity and protein levels. Other studies on the RTS pathway in T. cruzi have demonstrated the same stage-specific regulation of this pathway and have shown a likely association with the complex life cycle of this parasite and the availability of sulfur-containing amino acids in different parasite environments [33,34].
We found significantly lower levels of total thiol content in T. rangeli compared to T. cruzi epimastigotes. Based on the fact that cysteine forms the basic building block of all thiol antioxidants [2], one possible explanation for the lowest thiol levels observed may be because T. rangeli only uses the RTS pathway as a cysteine biosynthesis source. Another important aspect is related to the fact that exogenous organic sulfur-containing amino acids can be supplied by transporters [3,9,36]. However, such a mechanism and its possible influence on the total thiol levels in T. rangeli remain to be explored.
Different from T. cruzi, which faces oxidative stress in the mammalian host and within the triatomine vector’s digestive tract, T. rangeli is exposed to further oxidative and nitrosative stress while reaching the triatomine hemolymph and salivary glands [37]. Recently, studies have demonstrated the activation of the vector immune system during T. rangeli-Rhodnius prolixus interactions, including the generation of nitric oxide and superoxide free radicals [38-40]. The greater resistance of T. rangeli to SNAP compared to T. cruzi could be explained by the ability of T. rangeli to modulate insect immune/cellular factors [38,41], especially those related to nitrosative production, thus allowing the parasite to survive and multiply freely in the insect’s hemolymph and to invade and complete its development within the salivary glands [42].
Because thiols have been demonstrated to be the central metabolites in the redox metabolism of several parasite species [43], thus playing an important role in protection against oxidative stress, the higher T. rangeli susceptibility to hydrogen peroxide may be due its reduced total thiol content. In addition, the absence of an active CS enzyme potentiates the T. rangeli susceptibility to hydrogen peroxide, leading to the death of the parasite. Such findings are in agreement with reports in amoebae, whereby the overexpression of CS increases the total cellular thiol content and the resistance to oxidative stress due to hydrogen peroxide [8].
Conclusion
These findings demonstrate that the RTS pathway is active in T. rangeli, suggesting that this may be the only pathway for cysteine biosynthesis in this parasite because no CS activity was detected in epimastigotes and trypomastigotes and the CS genes are truncated due to the presence of stop codons. In this sense, the RTS pathway would have an important functional role during the insect stage of the life cycle of this protozoan parasite.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
IR and JT participated in the conception and design of the study and wrote the manuscript. LY was involved in cloning CβS. MS, AR, and EG were involved in the conception of the study and wrote the manuscript. All authors read and approved the final version of the manuscript.
Contributor Information
Ibeth Romero, Email: ibicris@gmail.com.
Jair Téllez, Email: jaircinio@gmail.com.
Lais Eiko Yamanaka, Email: laisyamanaka@gmail.com.
Mario Steindel, Email: mario.steindel@ufsc.br.
Alvaro José Romanha, Email: alvaro.romanha@ufsc.br.
Edmundo Carlos Grisard, Email: edmundo.grisard@ufsc.br.
Acknowledgements
IR, JT, and LY are recipients of CAPES or CNPq/PIBIC scholarships. This work was supported by CAPES, FINEP, and CNPq - Brazilian Government Agencies. The funders had no role in the study design, data generation and analysis, decision to publish, or preparation of the manuscript. The authors thank Dr Concepción Judith Puerta (Pontificia Universidad Javeriana, Bogota, Colombia) for critical reading and suggestions on the manuscript. We thank Aline Daiane Schlindwein, MSc for technical assistance in CβS and CS sequencing. IR and JT give a special acknowledgment in memory of John Walker, our friend and colleague.
References
- Nozaki T, Ali V, Tokoro M. Sulfur-containing amino acid metabolism in parasitic protozoa. Adv Parasitol. 2005;60:1–99. doi: 10.1016/S0065-308X(05)60001-2. [DOI] [PubMed] [Google Scholar]
- Krauth-Siegel RL, Leroux AE. Low-molecular-mass antioxidants in parasites. Antioxid Redox Signal. 2012;17(4):583–607. doi: 10.1089/ars.2011.4392. [DOI] [PubMed] [Google Scholar]
- Canepa GE, Bouvier LA, Miranda MR, Uttaro AD, Pereira CA. Characterization of Trypanosoma cruzi L-cysteine transport mechanisms and their adaptive regulation. FEMS Microbiol Lett. 2009;292(1):27–32. doi: 10.1111/j.1574-6968.2008.01467.x. [DOI] [PubMed] [Google Scholar]
- Beinert H, Holm RH, Münck E. Iron-sulfur clusters: nature’s modular, multipurpose structures. Science. 1997;277(5326):653–659. doi: 10.1126/science.277.5326.653. [DOI] [PubMed] [Google Scholar]
- Thomas D, Surdin-Kerjan Y. Metabolism of sulfur amino acids in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 1997;61(4):503–532. doi: 10.1128/mmbr.61.4.503-532.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aitken SM, Kirsch JF. The enzymology of cystathionine biosynthesis: strategies for the control of substrate and reaction specificity. Arch Biochem Biophys. 2005;433(1):166–175. doi: 10.1016/j.abb.2004.08.024. [DOI] [PubMed] [Google Scholar]
- Feldman-Salit A, Wirtz M, Hell R, Wade RC. A mechanistic model of the cysteine synthase complex. J Mol Biol. 2009;386(1):37–59. doi: 10.1016/j.jmb.2008.08.075. [DOI] [PubMed] [Google Scholar]
- Nozaki T, Asai T, Sanchez LB, Kobayashi S, Nakazawa M, Takeuchi T. Characterization of the gene encoding serine acetyltransferase, a regulated enzyme of cysteine biosynthesis from the protist parasites Entamoeba histolytica and Entamoeba dispar. Regulation and possible function of the cysteine biosynthetic pathway in Entamoeba. J Biol Chem. 1999;274(45):32445–32452. doi: 10.1074/jbc.274.45.32445. [DOI] [PubMed] [Google Scholar]
- Williams RA, Westrop GD, Coombs GH. Two pathways for cysteine biosynthesis in Leishmania major. Biochem J. 2009;420(3):451–462. doi: 10.1042/BJ20082441. [DOI] [PubMed] [Google Scholar]
- Raj I, Kumar S, Gourinath S. The narrow active-site cleft of O-acetylserine sulfhydrylase from Leishmania donovani allows complex formation with serine acetyltransferases with a range of C-terminal sequences. Acta Crystallogr D Biol Crystallogr. 2012;68(Pt 8):909–919. doi: 10.1107/S0907444912016459. [DOI] [PubMed] [Google Scholar]
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, Gocayne JD, Amanatides P, Ballew RM, Huson DH, Wortman JR, Zhang Q, Kodira CD, Zheng XH, Chen L, Skupski M, Subramanian G, Thomas PD, Zhang J, Gabor Miklos GL, Nelson C, Broder S, Clark AG, Nadeau J, McKusick VA, Zinder N. et al. The sequence of the human genome. Science. 2001;291(5507):1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- Wirtz M, Berkowitz O, Droux M, Hell R. The cysteine synthase complex from plants. Mitochondrial serine acetyltransferase from Arabidopsis thaliana carries a bifunctional domain for catalysis and protein-protein interaction. Eur J Biochem. 2001;268(3):686–693. doi: 10.1046/j.1432-1327.2001.01920.x. [DOI] [PubMed] [Google Scholar]
- Thomson L, Denicola A, Radi R. The trypanothione-thiol system in Trypanosoma cruzi as a key antioxidant mechanism against peroxynitrite-mediated cytotoxicity. Arch Biochem Biophys. 2003;412(1):55–64. doi: 10.1016/s0003-9861(02)00745-2. [DOI] [PubMed] [Google Scholar]
- Piacenza L, Zago MP, Peluffo G, Alvarez MN, Basombrio MA, Radi R. Enzymes of the antioxidant network as novel determiners of Trypanosoma cruzi virulence. Int J Parasitol. 2009;39(13):1455–1464. doi: 10.1016/j.ijpara.2009.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Alessandro A, Saraiva N. In: Parasitic Protozoa. 2. Kreirer JP BJ, editor. Vol. 2. San Diego: Academic Press; 1992. Trypanosoma rangeli; pp. 1–54. [Google Scholar]
- Camargo EP. Growth and differentiation in Trypanosoma cruzi I. Origin of metacyclic trypanosomes in liquid media. Rev Inst Med Trop Sao Paulo. 1964;6:93–100. [PubMed] [Google Scholar]
- Koerich LB, Emmanuelle-Machado P, Santos K, Grisard EC, Steindel M. Differentiation of Trypanosoma rangeli: high production of infective Trypomastigote forms in vitro. Parasitol Res. 2002;88(1):21–25. doi: 10.1007/s004360100501. [DOI] [PubMed] [Google Scholar]
- Pinto AM, Sales PC, Camargos ER, Silva AM. Tumour necrosis factor (TNF)-mediated NF-κB activation facilitates cellular invasion of non-professional phagocytic epithelial cell lines by Trypanosoma cruzi. Cell Microbiol. 2011;13:1518–1529. doi: 10.1111/j.1462-5822.2011.01636.x. [DOI] [PubMed] [Google Scholar]
- Grisard EC, Stoco PH, Wagner G, Sincero TC, Rotava G, Rodrigues JB, Snoeijer CQ, Koerich LB, Sperandio MM, Bayer-Santos E, Fragoso SP, Goldenberg S, Triana O, Vallejo GA, Tyler KM, Dávila AM, Steindel M. Transcriptomic analyses of the avirulent protozoan parasite Trypanosoma rangeli. Mol Biochem Parasitol. 2010;174(1):18–25. doi: 10.1016/j.molbiopara.2010.06.008. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. Molecular Cloning : a Laboratory Manual. 3. Cold Spring Harbor, N.Y.: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- Ewing B, Hillier L, Wendl MC, Green P. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 1998;8(3):175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- Decuypere S, Vanaerschot M, Brunker K, Imamura H, Müller S, Khanal B, Rijal S, Dujardin JC, Coombs GH. Molecular mechanisms of drug resistance in natural Leishmania populations vary with genetic background. PLoS Negl Trop Dis. 2012;6(2):e1514. doi: 10.1371/journal.pntd.0001514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher S, Winston SE, Fuller SA, Hurrell JG. Immunoblotting and immunodetection. Curr Protoc Mol Biol. 2008;Chapter 10 doi: 10.1002/0471142727.mb1008s83. Unit 10.18: 10.8.1-10.8.28. [DOI] [PubMed] [Google Scholar]
- Walker J, Barrett J. Cystathionine beta-synthase and gamma-cystathionase in helminths. Parasitol Res. 1991;77(8):709–713. doi: 10.1007/BF00928687. [DOI] [PubMed] [Google Scholar]
- Kashiwamata S, Greenberg DM. Studies on cystathionine synthase of rat liver. Properties of the highly purified enzyme. Biochim Biophys Acta. 1970;212(3):488–500. doi: 10.1016/0005-2744(70)90255-x. [DOI] [PubMed] [Google Scholar]
- Gaitonde MK. A spectrophotometric method for the direct determination of cysteine in the presence of other naturally occurring amino acids. Biochem J. 1967;104(2):627–633. doi: 10.1042/bj1040627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Droux M, Martin J, Sajus P, Douce R. Purification and characterization of O-acetylserine (thiol) lyase from spinach chloroplasts. Arch Biochem Biophys. 1992;295(2):379–390. doi: 10.1016/0003-9861(92)90531-z. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay R, Dey S, Xu N, Gage D, Lightbody J, Ouellette M, Rosen BP. Trypanothione overproduction and resistance to antimonials and arsenicals in Leishmania. Proc Natl Acad Sci U S A. 1996;93(19):10383–10387. doi: 10.1073/pnas.93.19.10383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellman GL. Tissue sulfhydryl groups. Arch Biochem Biophys. 1959;82(1):70–77. doi: 10.1016/0003-9861(59)90090-6. [DOI] [PubMed] [Google Scholar]
- Räz B, Iten M, Grether-Bühler Y, Kaminsky R, Brun R. The Alamar Blue assay to determine drug sensitivity of African trypanosomes (T.b. rhodesiense and T.b. gambiense) in vitro. Acta Trop. 1997;68(2):139–147. doi: 10.1016/s0001-706x(97)00079-x. [DOI] [PubMed] [Google Scholar]
- Kelly JM, Taylor MC, Smith K, Hunter KJ, Fairlamb AH. Phenotype of recombinant Leishmania donovani and Trypanosoma cruzi which over-express trypanothione reductase. Sensitivity towards agents that are thought to induce oxidative stress. Eur J Biochem. 1993;218(1):29–37. doi: 10.1111/j.1432-1033.1993.tb18348.x. [DOI] [PubMed] [Google Scholar]
- Taoka S, Widjaja L, Banerjee R. Assignment of enzymatic functions to specific regions of the PLP-dependent heme protein cystathionine beta-synthase. Biochemistry. 1999;38(40):13155–13161. doi: 10.1021/bi990865t. [DOI] [PubMed] [Google Scholar]
- Nozaki T, Shigeta Y, Saito-Nakano Y, Imada M, Kruger WD. Characterization of transsulfuration and cysteine biosynthetic pathways in the protozoan hemoflagellate, Trypanosoma cruzi. Isolation and molecular characterization of cystathionine beta-synthase and serine acetyltransferase from Trypanosoma. J Biol Chem. 2001;276(9):6516–6523. doi: 10.1074/jbc.M009774200. [DOI] [PubMed] [Google Scholar]
- Marciano D, Santana M, Nowicki C. Functional characterization of enzymes involved in cysteine biosynthesis and H(2)S production in Trypanosoma cruzi. Mol Biochem Parasitol. 2012;185(2):114–120. doi: 10.1016/j.molbiopara.2012.07.009. [DOI] [PubMed] [Google Scholar]
- Yarlett N, Bacchi CJ. Effect of DL-alpha-difluoromethylornithine on methionine cycle intermediates in Trypanosoma brucei brucei. Mol Biochem Parasitol. 1988;27(1):1–10. doi: 10.1016/0166-6851(88)90019-9. [DOI] [PubMed] [Google Scholar]
- Duszenko M, Ferguson MA, Lamont GS, Rifkin MR, Cross GA. Cysteine eliminates the feeder cell requirement for cultivation of Trypanosoma brucei bloodstream forms in vitro. J Exp Med. 1985;162(4):1256–1263. doi: 10.1084/jem.162.4.1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azambuja P, Ratcliffe NA, Garcia ES. Towards an understanding of the interactions of Trypanosoma cruzi and Trypanosoma rangeli within the reduviid insect host Rhodnius prolixus. An Acad Bras Cienc. 2005;77(3):397–404. doi: 10.1590/s0001-37652005000300004. [DOI] [PubMed] [Google Scholar]
- Whitten MM, Mello CB, Gomes SA, Nigam Y, Azambuja P, Garcia ES, Ratcliffe NA. Role of superoxide and reactive nitrogen intermediates in Rhodnius prolixus (Reduviidae)/Trypanosoma rangeli interactions. Exp Parasitol. 2001;98(1):44–57. doi: 10.1006/expr.2001.4615. [DOI] [PubMed] [Google Scholar]
- Whitten M, Sun F, Tew I, Schaub G, Soukou C, Nappi A, Ratcliffe N. Differential modulation of Rhodnius prolixus nitric oxide activities following challenge with Trypanosoma rangeli, T. cruzi and bacterial cell wall components. Insect Biochem Mol Biol. 2007;37(5):440–452. doi: 10.1016/j.ibmb.2007.02.001. [DOI] [PubMed] [Google Scholar]
- Garcia ES, Castro DP, Figueiredo MB, Genta FA, Azambuja P. Trypanosoma rangeli: a new perspective for studying the modulation of immune reactions of Rhodnius prolixus. Parasit Vectors. 2009;2(1):33. doi: 10.1186/1756-3305-2-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazos-Lopes F, Mesquita RD, Silva-Cardoso L, Senna R, Silveira AB, Jablonka W, Cudischevitch CO, Carneiro AB, Machado EA, Lima LG, Monteiro RQ, Nussenzveig RH, Folly E, Romeiro A, Vanbeselaere J, Mendonça-Previato L, Previato JO, Valenzuela JG, Ribeiro JM, Atella GC, Silva-Neto MA. Glycoinositolphospholipids from Trypanosomatids subvert nitric oxide production in Rhodnius prolixus salivary glands. PLoS One. 2012;7(10):e47285. doi: 10.1371/journal.pone.0047285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia ES, Castro DP, Figueiredo MB, Azambuja P. Parasite-mediated interactions within the insect vector: trypanosoma rangeli strategies. Parasit Vectors. 2012;5:105. doi: 10.1186/1756-3305-5-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krauth-Siegel RL, Comini MA. Redox control in trypanosomatids, parasitic protozoa with trypanothione-based thiol metabolism. Biochim Biophys Acta. 2008;1780(11):1236–1248. doi: 10.1016/j.bbagen.2008.03.006. [DOI] [PubMed] [Google Scholar]