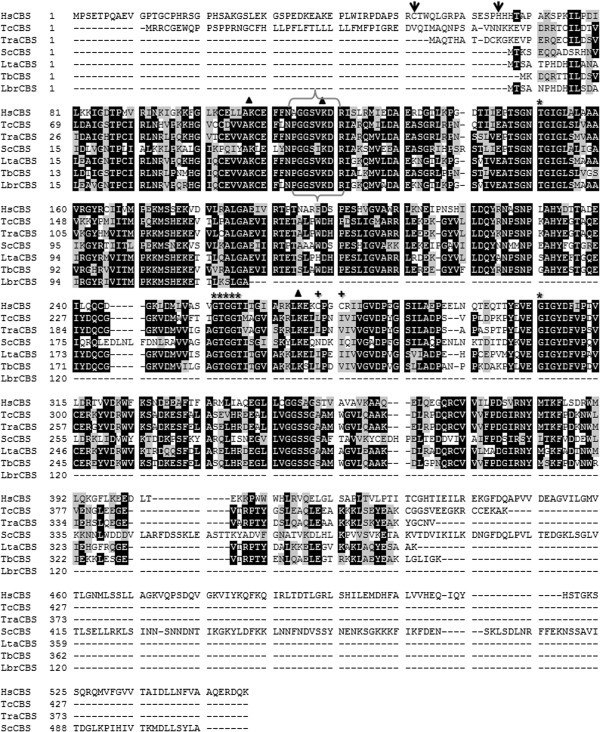
Figure 1.
Multiple alignment of deduced amino acid sequences of CβS from T. rangeli (TrCβS) and other representative organisms. The identity (black background) and conservation (grey background) of the amino acid residues are shown. Brackets indicate the consensus amino acid residues of the putative pyridoxal phosphate-binding motif (PXXSVKDR), and other motifs vital for CβS activity are indicated with asterisks (*). The oxido-reductase motif of HsCβS is highlighted with (+). The lysine residues required for CS catalytic activity are marked with triangles. The positions of the heme-binding residues within the heme domain of the human CβS enzyme (Cys52 and His65) are marked with (↓). HsCβS: Human (P35520); TcCβS: Trypanosoma cruzi (Tc00.1047053511691.20); ScCβS: Saccharomyces cerevisiae (P32582) LtaCβS: Leishmania tarentolae (LtaP17.0270); TbCβS: Trypanosoma brucei (Tb11.02.5400); LbrCβS: Leishmania braziliensis (LbrM.17.0230).