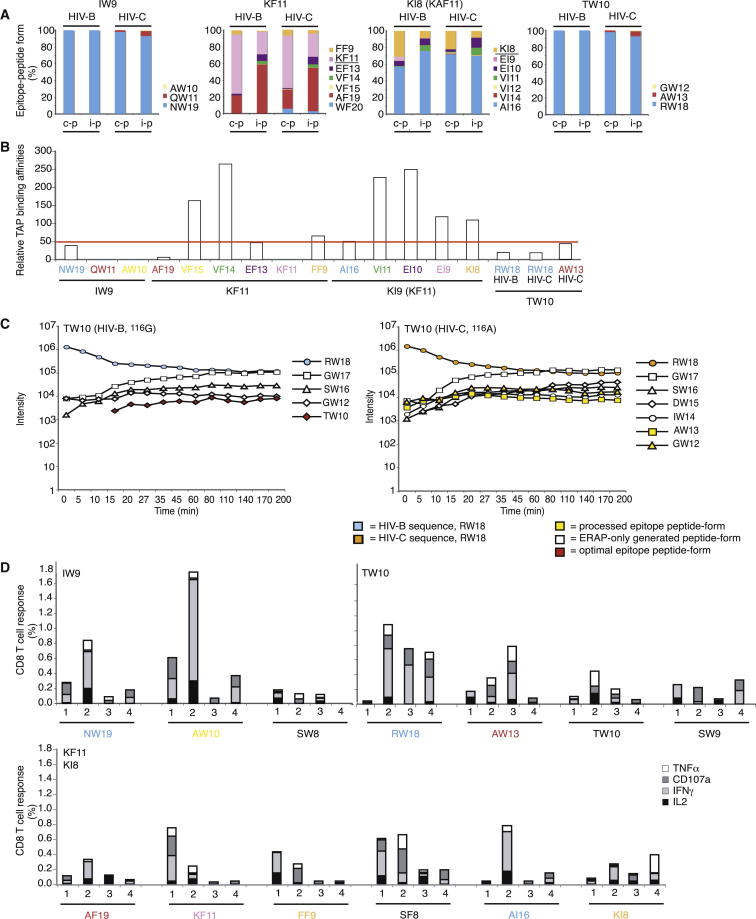
Figure 2.
TAP Binding, ERAP Trimming, and CD8+ T Cell Analyses
(A) Overview of the abundance of all IW9, KF11, KI8, and TW10 epitope forms produced after 4 hr constitutive (c-p) or immunoproteasomal (i-p) digestion.
(B) TAP binding affinity of the IW9, KF11, KI8, and TW10 epitope forms generated by proteasomal digestion. GW12 could not be tested because it was insoluble and WF20, VI14, and VI12 were not tested. The epitope forms are colored as in (A). The results represent one of at least two independent experiments.
(C) ERAP1,2 digestion of the TW10 epitope precursor RW18 from HIV B and HIV C. The result represents one of three independent experiments.
(D) PBMCs from four untreated HIV-1-infected patients with HLA-B∗5701 (Table S3) were stimulated with a subset of IW9, KF11, KI8, and TW10 HIV B epitope-precursor peptides because of limited sample availability. IL-2, IFN-γ, TNF-α, and CD107a responses were analyzed using flow cytometry. The percentages of responding CD8+ T cells are shown after background subtraction, and each peptide was tested two to three times. The epitope-peptide form is indicated along the x axis, and the optimal epitope is shown in the top-left corner of the graph. The epitope forms made by the proteasome are colored as in (A) and (B).