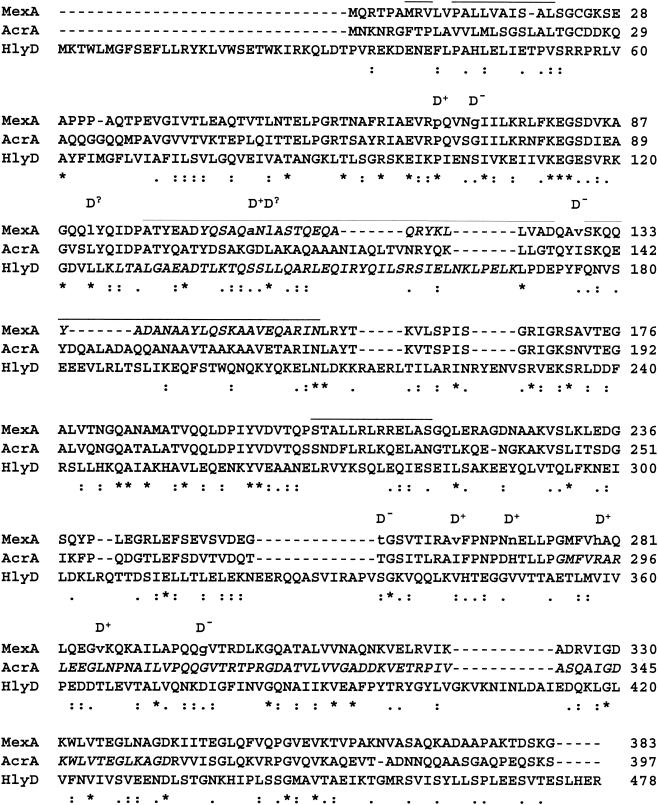
FIG. 5.
Multiple alignment of MFPs. The MexA, AcrA, and HlyD (accession number AAA23977) proteins were aligned on-line by using ClustalW (http://www.ebi.ac.uk/clustalw/) (78). Residues in MexA whose mutation in this study compromised MexA function are indicated in lowercase, and the impact of the mutation on MexA-MexA interaction is indicated above the residue (D−, dimerization deficient; D+, dimerization proficient; D?, dimerization could not be assessed because the mutant protein was not expressed in the E. coli SU101 strain used to assess MexA dimerization). Regions of MexA predicted to form α-helices (overlined) were identified by using programs available on-line from the JPred server (http://www.compbio.dundee.ac.uk/∼www-jpred/) (9), and two of these (shown in italics) were predicted by Johnson and Church (30) to form coiled-coil structures. The N-terminal region of HlyD implicated in promoting an interaction of HlyD with the OMF TolC (69) is indicated in italics. The C-terminal region of AcrA predicted to be involved in the interaction of this MFP with its cognate RND component, AcrB (16), is also indicated in italics.