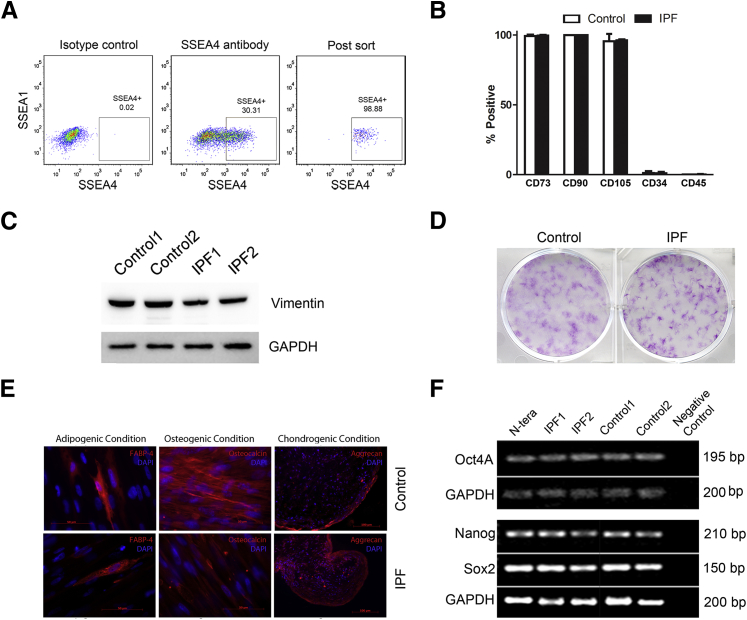
Figure 1.
Recovery and characterization of SSEA4+/SSEA1−/small cells from IPF and control primary mesenchymal cell lines. A: Representative of SSEA4+/SSEA1−/small cells that had undergone FACS. Primary mesenchymal cell lines were sorted by FACS to isolate cells with relatively high surface expression of SSEA4 and low surface expression of SSEA1. Shown are the gates used (solid lines) to define the SSEA4+/SSEA1− cell population. The boxed area indicates the region defined as SSEA4+/SSEA4−. Cells were also sorted on the basis of size (forward and side scatter were calibrated with cells passing through a 12-μm mesh). B: Flow cytometric analysis of SSEA4+/SSEA1−/small cells for expression of cell surface determinants used to define MSCs (CD73, CD90, CD105) and fibrocytes (CD34 and CD45). Data shown are from SSEA4+/SSEA1−/small cells from two control and two IPF primary mesenchymal cell lines (means ± SD). C: SSEA+/SSEA1−/small cells were examined for vimentin expression by Western blot analysis. GAPDH is shown as a loading control. D: Colony-forming unit fibroblast assay: SSEA4+/SSEA1−/small cells were seeded at clonal density onto plastic tissue culture dishes (500 cells/9.5 cm2) and maintained in culture for 14 days. Shown are representative images of crystal violet-stained colonies formed by SSEA4+/SSEA1−/small cells sorted from two control and two IPF primary mesenchymal cell lines, each assayed in triplicate. E: SSEA4+/SSEA1−/small cells were investigated for tri-lineage differentiation capacity: i) adipocytes as determined by immunoreactivity to anti-FABP4 and the presence of lipid within the cells; ii) osteocytes by immunoreactivity to anti-osteocalcin; and iii) chondrocytes, as determined by the presence of aggrecan. Data shown are representative of SSEA4+/SSEA1−/small cells sorted from three control and three IPF primary mesenchymal cell lines. F: RT-PCR analysis of IPF and control SSEA4+/SSEA1−/small cells for progenitor transcription factors: Oct4A, Nanog, and Sox2, with GAPDH serving as a loading control. Human testicular embryonal carcinoma cells (N-TERA) were used as a positive control and PCR with primers but no cDNA as a negative control. Data shown are representative of SSEA4+/SSEA1−/small cells sorted from four IPF and four control primary mesenchymal cell lines. The Oct4A band was sequenced from one IPF and one control to verify its identity. Scale bars: 50 μm (E, left and middle panels); 100 μm (E, right panels). FABP4, fatty acid binding protein 4; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.