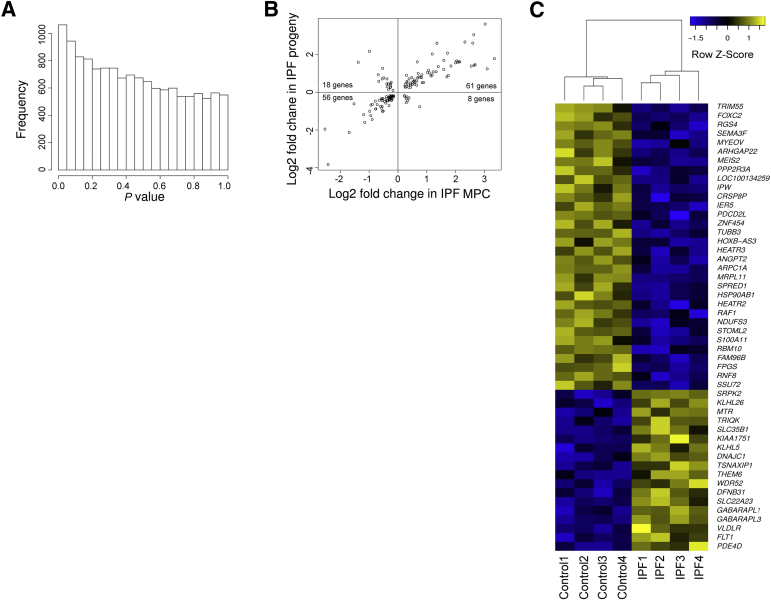
Figure 6.
Genes and pathways that distinguish IPF mesenchymal progenitor cells and their progeny from their control counterparts are co-regulated. Steady state mRNA profiles identify activation of cellular processes in IPF progeny consistent with their fibrotic phenotype. A: Histogram of gene-by-gene P values that compare IPF and control progeny. Data shown are from progeny derived from eight independent mesenchymal progenitor cell lines. B: Shown are log2 fold changes of genes altered in both IPF mesenchymal progenitors and IPF progeny. Co-regulated genes appear in the right upper and left lower quadrants of the plot. C: Heatmap of the 50 most significantly altered genes between IPF and control progeny. Colors represent per gene z-score (expression difference normalized for SD). Co-regulation of genes distinguishing IPF mesenchymal progenitor cells and IPF progeny from their control counterparts. P = 1.9 × 10−15, Fisher exact test (B). n = 4 (A, IPF and control).