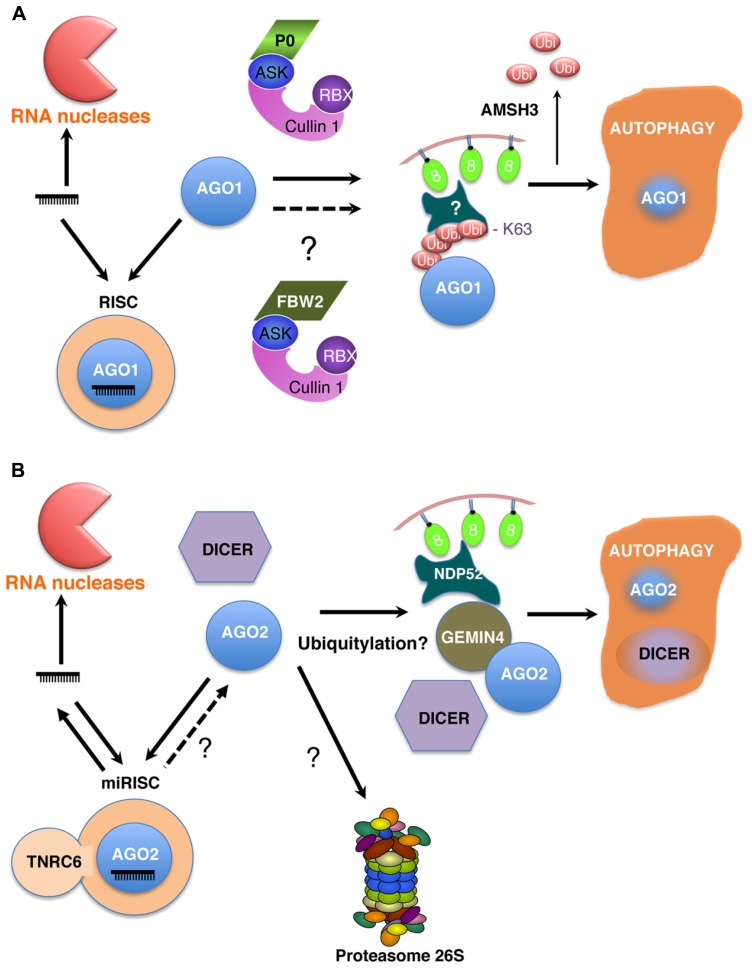
FIGURE 1.
Models for the turnover of AGO proteins in Arabidopsis (A) and mammalian cells (B). Different levels of regulation operate on the homeostasis of RISCs. First, the steady-state levels of microRNAs are regulated by degradation processes involving different ribonucleases (RNases) acting either 3′–5′ or 5′–3′. Thus microRNAs most likely are in competition for AGO binding. Recent evidences essentially from metazoans indicate that at least some microRNAs can be released from RISCs, explaining their short half-lives. However, not only microRNAs but also AGO proteins are degraded. Thus in both plants and animal cells, it is now clearly established that AGO proteins are degraded by autophagy in an RNA free form prior RISC assembly. This mechanism also co-degrades other components of the silencing machinery such as DICER in mammals (B). In Arabidopsis, the polerovirus protein P0 assembles an SCFP0 ubiquitin ligase to ubiquitylate AGO1 or an AGO1 associated protein (A). Viral P0-mediated AGO1 degradation by autophagy also requires the deubiquitylating enzyme AMSH3. However, the identity of endogenous ubiquitin ligases involved in this process have not yet been unambiguously identified. The role of ubiquitylation in the turnover of human AGO2 is at present unclear but requires GEMIN4 and NDP52 (B). Finally, whether upon guide RNA dissociation AGO2 would become accessible to autophagy and the role of the proteasome in AGOs degradation are other still open questions.