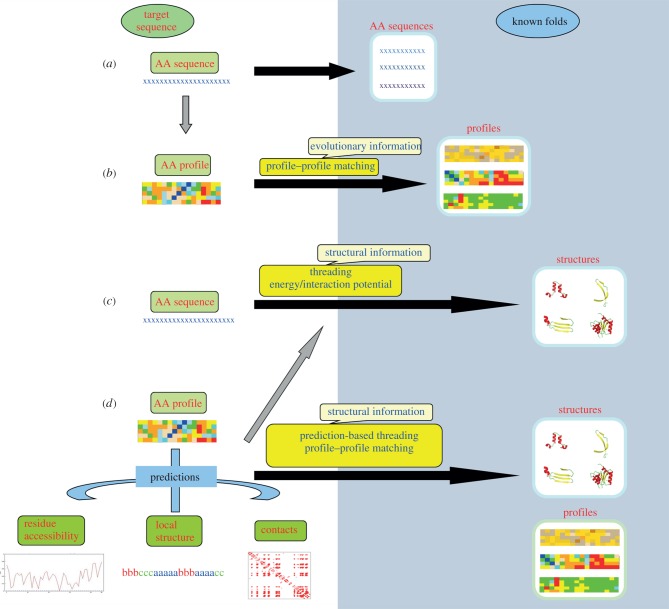
Figure 2.
Different strategies for protein fold recognition. The fold space is highlighted by the blue background and the lengths of the black arrows joining the target sequence (space) and fold space give an idea of the distance of relationship. (a) Close relationships are often detected by simple sequence alignment techniques. (b) Addition of evolutionary information using sequence profiles derived from MSAs helps in detecting more distantly related folds. When the sequence-based alignments are not informative, sequence–structure matching needs to be carried out. (c) The target sequence can be threaded on to the known folds to check the compatibility. The compatibility is usually quantified based on the global interaction or energy potential. Obtaining an optimal alignment between the sequence and a fold is however difficult and computationally quite expensive. (d) The other alternative is to carry out prediction of different structural features like local backbone conformation, solvent accessibility or contact order and then matching the predicted features with that found in the known fold. (Online version in colour.)