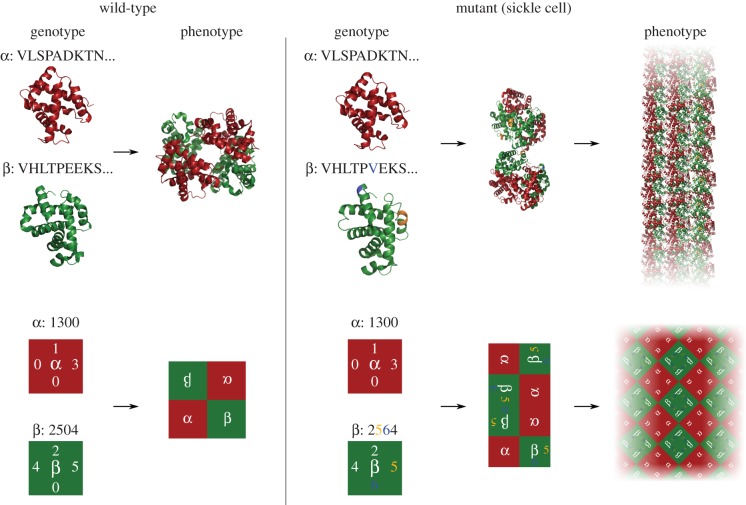
Figure 2.
The haemoglobin phenotype polymorphism with a representation in a Polyomino GP map. A Polyomino genotype (bottom left) is constructed to mimic the transition from the haemoglobin wild-type (top left, Hb A, PDB: IGZX [25]) to the sickle cell mutant (top right, Hb S, PDB: 2HBS [26]). In Hb S, residue 6 of the β chain is mutated from glutamic acid (E) to valine (V), shown in blue. The mutation leads to a hydrophobic patch being created at this position which allows binding to a pocket between residues 85 and 88 (shaded orange) on the second β chain. A single binding is depicted in the middle stage of the top right. The binding allows for large-scale polymerization of Hb S in a double strand form, which further aggregates to form extended fibres [27] (a cartoon depiction of this in the top right). In the polyomino example (bottom), we assume that odd-even numbered interfaces may interact (full details of the model are explained in §2). In the bottom right, a single change (from 0 to a 6 at the third residue of the β tile) has occurred resulting in a new interaction with the second position also on the β tile (5 ↔ 6). The new interaction leads the self-assembly process to produce an unbound aggregate, tiling the two-dimensional plane. This provides an example of the potential for the Polyomino GP map to capture basic features of protein assembly, including extended structures.