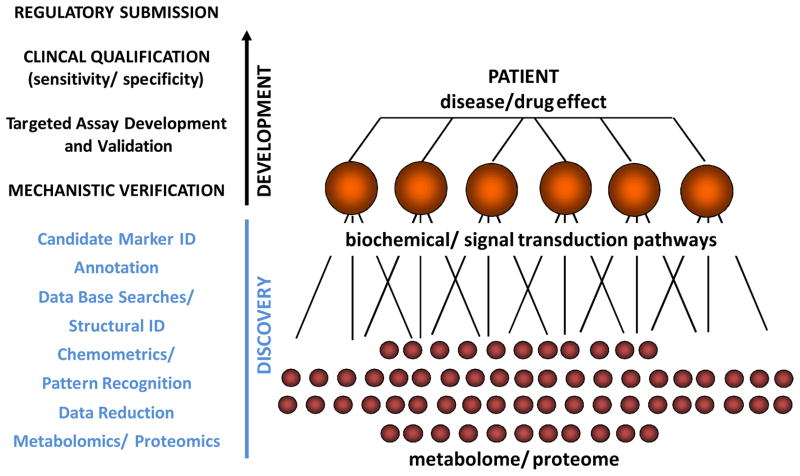
Figure 1. Steps During Discovery and Development of Molecular Markers.
Typically discovery involves the non-targeted analysis of the metabolome and/or proteome of the samples. This may result in hundreds and sometimes thousands of data points. The next step is to extract relevant data and to identify candidate surrogate markers of a disease process or drug effect. Making sense of the data is just as important as the bioanalytics and may be even more challenging and time consuming. Chemometrics is defined as the application of mathematical and statistical methods to chemistry. Chemometric analyses are necessary in order to develop statistical pattern recognition models, achieve optimal characterization of the samples and detect biomarkers from diverse, highly dimensional omics datasets [38]. After based on these analyses markers have been selected and their structures have been identified, it is necessary to assess if the markers truly are surrogates for the effect of a disease or drug. Annotation seeks to link the changes in molecular markers to biochemical and cell regulatory pathways. Verification is to establish a mechanistic link between specific markers and a disease process or the pharmacodynamic/toxicodynamic drug effects. Once the candidate markers have been mechanistically verified, their clinical utility in terms of their sensitivity and specificity to detect and/or predict a disease process, clinical outcome or drug response needs to be established (qualification). For this purpose it is often necessary to establish validated bioanalytical assays. Clinical qualification often requires the conduct of prospective multi-center clinical trials. If qualification is successful and a molecular marker shows good specificity and sensitivity, regulatory submission and approval may be required in most cases before the molecular marker can widely be used as a clinical diagnostic tool [64].