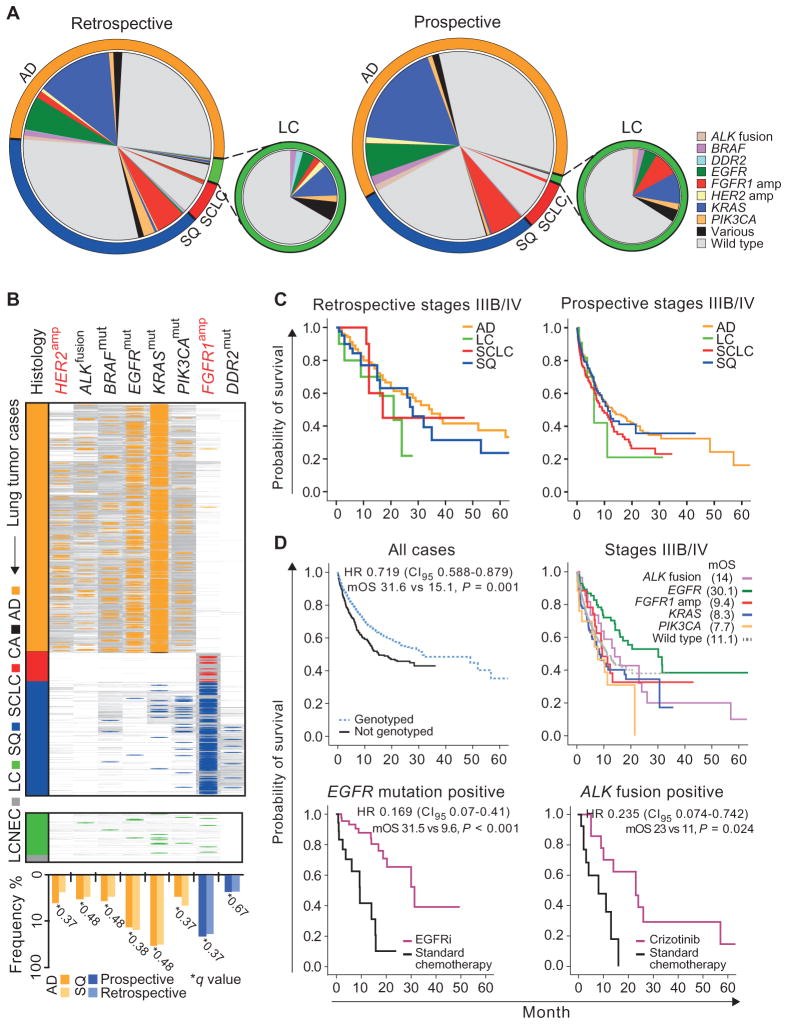
Fig. 5. Clinically relevant genome alterations in lung cancer subtypes.
(A) Genetic alterations per histological subtype in retrospective and prospective sample sets. Each chart represents the overall population with proportions of histological subtypes color-coded in the outer ring. Frequencies of alterations (wild type: no alteration in ALK, EGFR, FGFR1, KRAS, or PIK3CA) are given per gene relative to all cases within each histological subtype. Distribution of alterations for the LC population is shown separately. (B) Genotyping results of 3590 patients enrolled in the prospective screening effort are sorted according to the histological subtype [AD (2250), CA (3), SCLC (265), SQ (1018), LC (47), and LCNEC (7)]. Colored lines indicate alterations, and gray lines indicate wild type. Frequencies of alterations for AD (orange) and SQ (blue) are plotted below the respective genes, comparing the mutation frequency in the prospective data set (dark colors) to the retrospective data set (light colors). No significant difference between the data sets was seen (q values are given on the graph; P values were adjusted for multiple hypothesis testing using the Benjamini and Hochberg method). Color code: orange, AD; black, CA; green, LC; gray, LCNEC; red, SCLC; blue, SQ. (C) Kaplan-Meier curves for overall survival are shown for stage IIIB/IV patients per histological subtype for the retrospective (left) and prospective (right) sample sets. No significant difference was seen between subtypes within each data set. (D) Prospective sample set: Kaplan-Meier curves for overall survival are shown for all patients who were genetically tested versus those without available genetic information (top left). Overall survival is shown for stage IIIB/IV patients with alterations in given genes versus patients with wild type in the given genes (top right). Overall survival was statistically significantly longer in EGFR-mutant cases compared to all other (log-rank test, P < 0.05) except ALK-rearranged cases (P = 0.065). Overall survival is shown for patients with EGFR mutation treated with an EGFR inhibitor or standard chemotherapy (bottom left) and patients with ALK translocations treated with crizotinib or standard chemotherapy (bottom right). Gain of overall survival in the patient group treated with kinase inhibitors versus standard chemotherapy is given by the median overall survival (mOS). P values were corrected using the Bonferroni adjustment. HR, hazard ratio.