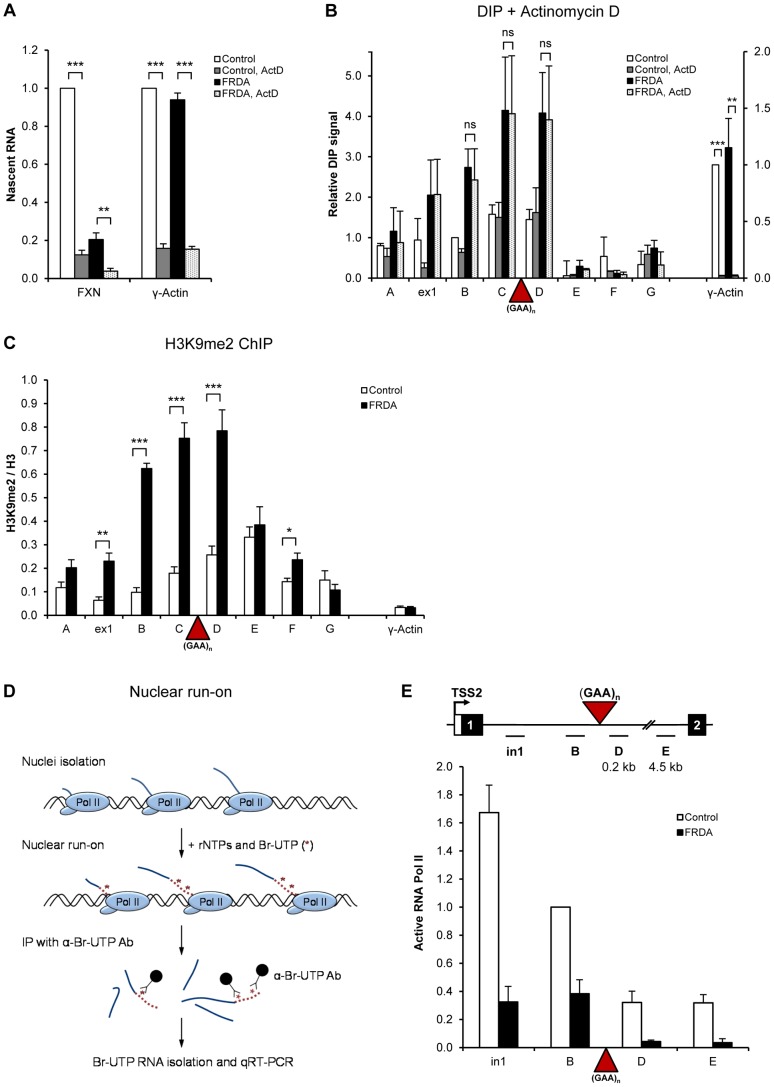
Figure 2. R-loops are stable and impede Pol II transcription on FXN gene.
A. RT-qPCR analysis of nascent γ-actin and FXN RNA from control and FRDA cells treated with 5 µg/ml of actinomycin D for 21 hours. Values are relative to untreated control cells. B. DIP on FXN gene in control and FRDA cells treated with 5 µg/ml of actinomycin D for 21 hours. γ-actin is positive control. C. H3K9me2 ChIP in control and FRDA cells. H3K9me2 levels were normalized to the total H3 levels. γ-actin is used as background control. D. Diagram depicting the Br-UTP nuclear run-on (NRO) method. E. Br-UTP nuclear run-on in two control (GM15851, GM14926) and two FRDA (GM15850 and GM16243) cells, normalised to the region B in control cells. Bars in A–C and E are average values +/− SEM (n>3).