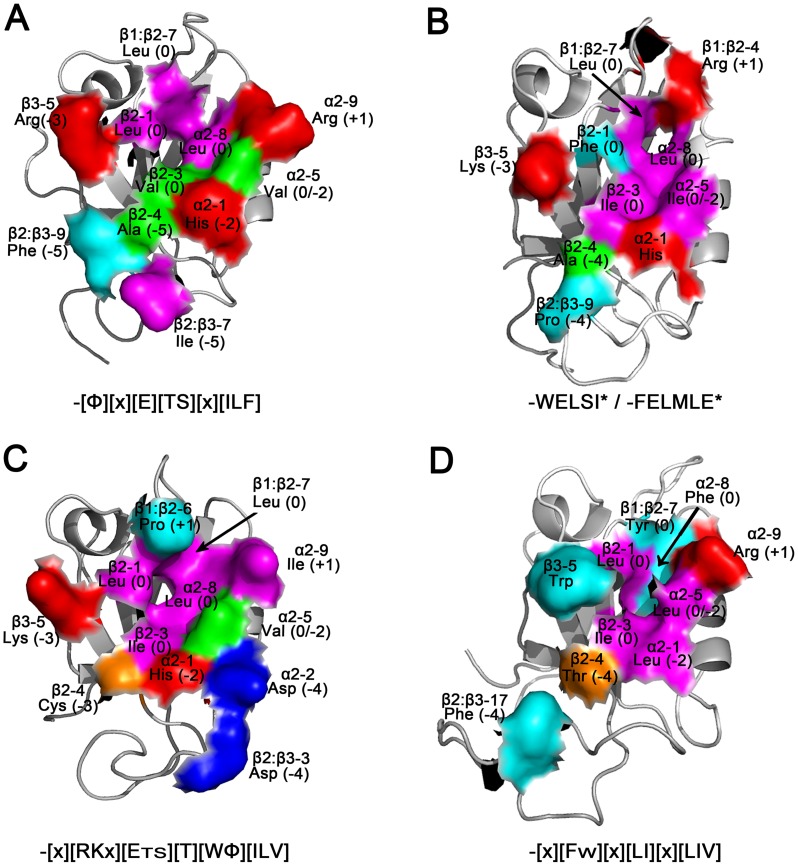
Figure 6. Modeling of ligand binding specificities for the SjScrib-PDZ domains.
The predicted structures of the first (A), second (B), third (C), and fourth (D) PDZ domain of SjScrib are shown and colored to highlight the functional residues (shown as a partial molecular surface model) involved in ligand recognition. The position in an element of secondary, name, and the potential ligand subsite involved in recognition for each key residue are denoted. Color schemes: large hydrophobic residues (magenta); small hydrophobic residues (green); small polar residues (orange); aromatic residues (cyan); positively charged residues (red); negatively charged residues (blue). The ligand specificities for each PDZ domain are shown under each panel. Structural figures were produced with PyMOL.