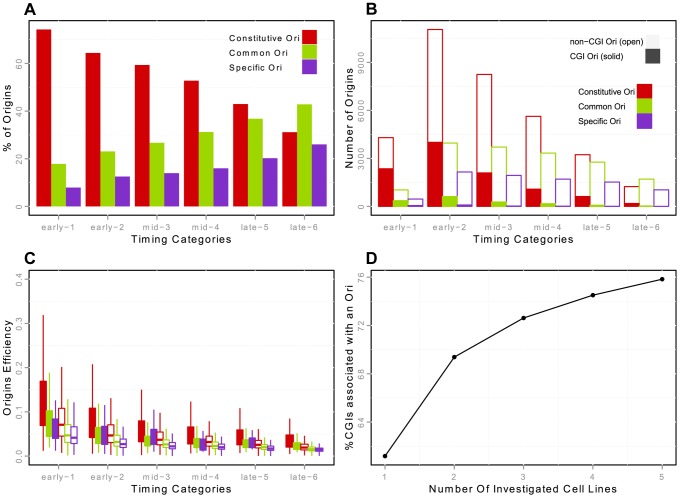
Figure 2. Constitutive, common and cell specific replication origins (K562) and association with CGIs.
A: Percentage of constitutive/common/K562-specific origins in each timing category (from early to late origins, as explained in the Methods Section). Constitutive origins are determined by the intersection of the origins from the five cell lines -H9, HeLa, IMR90, K562 and iPS- with Galaxy [47]. B: Number of constitutive/common/K562-specific origins in each timing category, with distinction between origins associated and not associated with CGIs. Origins classified as CGIs correspond to origins strictly overlapping a CGI. Positions of CGIs are taken from UCSC Genome Browser annotation. C: Boxplots of origin efficiency according to timing, constitutive/common/K562-specific nature and association with CGIs. Origin efficiency is defined as the number of reads within the origin interval divided by the length of the origin (i.e. the density of reads within a given origin). D: Proportion of CGIs overlapping with an origin in at least one cell line, according to the number of cell lines analyzed.