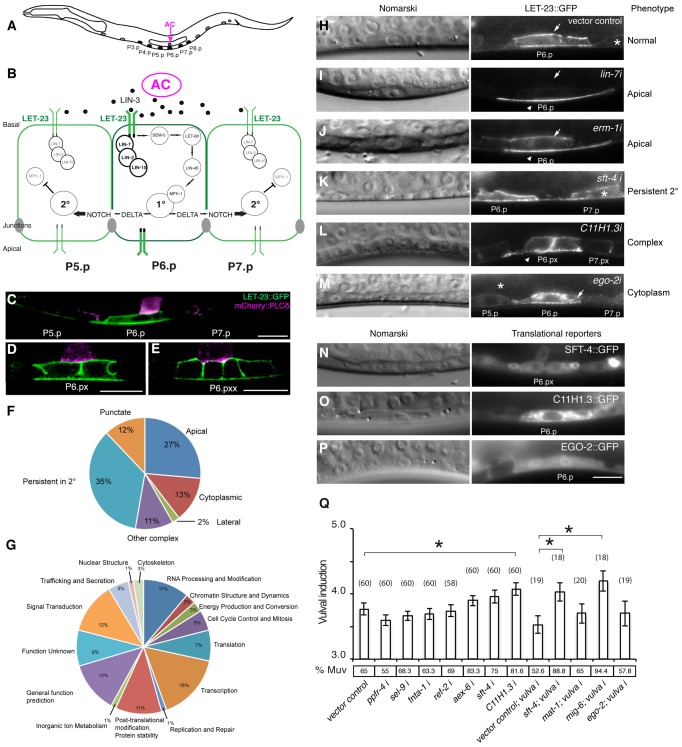
Figure 1. Identification of genes regulating LET-23 EGFR localization and signaling.
(A) Schematic drawing of an L2 larva with the location of the VPCs and AC. P5.p, P6.p and P7.p get induced to form the mature vulva. P3.p, P4.p and P8.p divide once and fuse to the hypodermis. (B) Overview of the LET-23 EGFR and NOTCH signaling network controlling 1° and 2° vulval fate specification. (C) LET-23::GFP expression (green) in P6.p of a late L2 larva during vulval induction. The AC is labeled with an mCherry::plcδPH reporter (magenta) [35]. Note the low LET-23::GFP levels in the 2° P5.p and P7.p. (D) Expression of the LET-23::GFP reporter in the 1° lineage at the Pn.px and (E) Pn.pxx stage. (F) Pie charts indicating the frequencies of the different classes of mislocalization phenotypes observed after RNAi and (G) the Clusters of Orthologous Groups (KOGs) of the 81 genes identified in the screen (H–M). Examples of different genes identified in the LET-23 localization screen. Left panels show the corresponding Nomarski images and right panels LET-23::GFP expression in the 1° cells and their neighbors (asterisks). (H) The negative empty vector control and (I) lin-7 RNAi as positive control. (J) erm-1 RNAi as an example for reduced basolateral (arrow) and increased apical localization (arrow head), (K) sft-4 RNAi with normal localization in P6.p but persistent expression in P7.p (asterisk), and (L) C11H1.3 RNAi (Pn.px stage) with punctate apical accumulation (arrow head). (M) ego-2 RNAi with cytoplasmic accumulation of LET-23::GFP in P6.p (arrow head) and P5.p (asterisk). (N) perinuclear localization of SFT-4::GFP in the vulval cells and the AC and (O) intracellular punctate expression of C11H1.3::GFP in P6.p. (P) Cytoplasmic and nuclear expression of EGO-2::GFP in P6.p. (Q) Vulval induction in let-60(n1046gf) larvae treated with different RNAi clones. Vulval induction (VI) indicates the average number of induced VPCs per animal. “vulva i” indicates Pn.p cell-specific RNAi in the rde-1(lf);let-60(n1046gf); [Plin-31::rde-1] background. %Muv (Multivulva) indicates the fraction of animals with VI>3. The numbers of animals scored are indicated in brackets. * Indicates p<0.05 as determined in a two tailed student's t-test - two-sample unequal variance. t-test values in RNAi: C11H1.3 (0.003), sft-4 (0.013), mig-6 (0.002). Error bars represent the standard error of the mean. The scale bars are 10 µm.