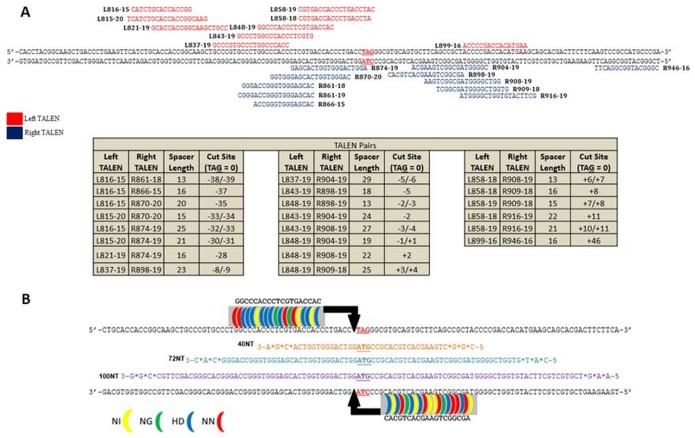
Figure 2. Array of TALENs Designed for Proximal Cleavage Analysis.
(A) Nine Left and eleven Right TALEN half-sites were designed to flank the target at a range of −39 to +46 base pairs of the integrated mutant eGFP gene (TAG = 0). TALENs were designed according to previously published guidelines (see materials and methods). The 20 constructed TALEN half-sites, 9 left depicted in red, and 11 right depicted in blue, were combined for targeting experiments. By using all possible combinations of TALEN half-sites with spacers between 13–29 bases; 22 total TALEN combinations were tested that flank the mutant base. Cut site reflects the predicted position of ds break induced by the TALEN pair relative to the mutant base (TAG). (B) The TALEN pair, designed and built using the Golden Gate method, induces a double stranded break immediately preceding the mutant codon. RVDs are shown as color coded binding blocks next to their respective base, yellow NI:A, green NG:T, blue HD:C and red NN:G. Fok1 domains are shown in black and are positioned at their predicted cut site.