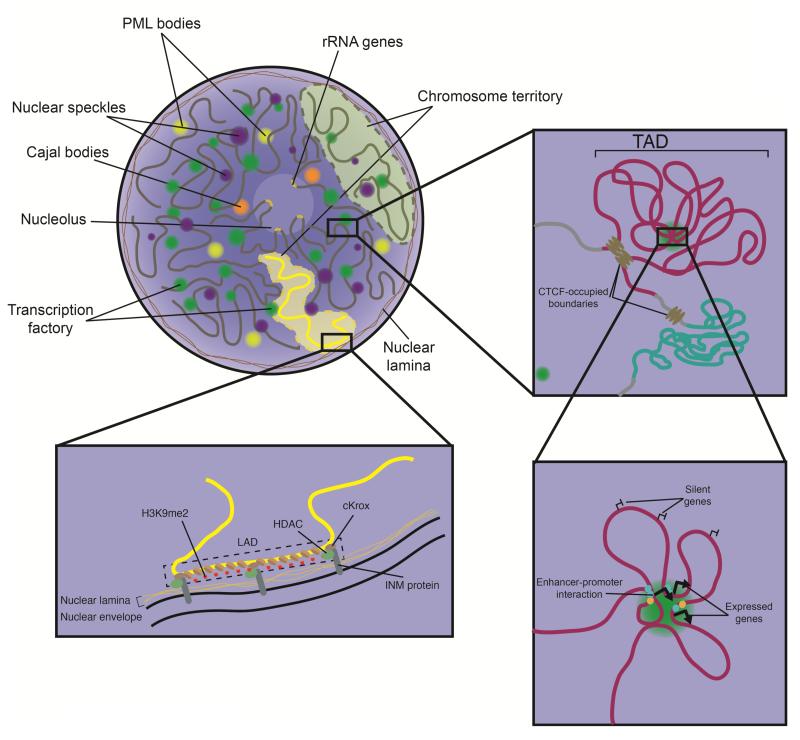
Figure 1.
Organization of chromatin in topological and lamina-associated domains. A. Cartoon model for the higher-order organization of chromatin. A mammalian nucleus shows chromosomes (dark grey lines) confined to distinct territories. rRNA genes are confined within the nucleolus. Cajal bodies are shown juxtaposed to the nucleolus. Nuclear speckles and transcription factories reflect high concentrations of the splicing and transcriptional machinery, respectively. These structures are depleted in nuclear compartments associated with repressed heterochromatin: the nuclear lamina and chromatin associated with the nucleolus. On right, a magnified view of the boxed region shows two topological associated domains (TADs), colored red and green, separated by CTCF-bound boundary regions. Within each TAD are numerous chromosomal interactions; however, few interactions cross boundaries between TADs. On bottom right, further magnification of a transcription factory with a TAD demonstrates close associations between distal regulatory regions and expressed genes. Moreover, numerous expressed genes colocalize in this space and share similar sets of transcription factors (green and orange circles). Silent genes found in between these active genes in linear sequence loop away from the transcription factory and occupy a distinct region. On bottom left, a repressed chromatin domain is associated with the nuclear lamina, known as a lamina-associated domain (LADs). LADs have hallmarks that include high levels of repressive H3K9 methylation. Interactions between inner nuclear membrane proteins, such as emerin and Lap2β, with HDACs and cKrox are also essential for LAD establishment.