Figure 4. Spd1 and PCNA interact in vivo and a PIP box in Spd1 is important for this interaction.
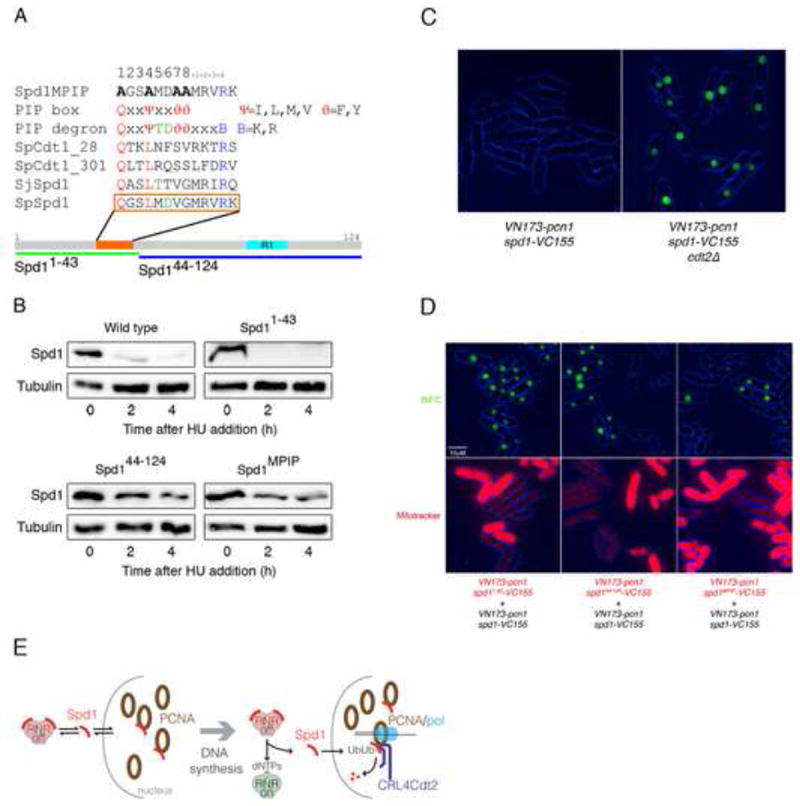
(A) Identification of a sequence (orange bar and box) in Spd1 (grey bar) that partially matches the PIP degron consensus sequence. Other sequences shown are (bottom to top): corresponding region of S. japonicus Spd1 (SjSpd1); PIP degrons in S. pombe Cdt1 (SpCdt1_301, SpCdt1_28 [18]): PIP degron and PIP consensus sequences [30]; and the mutated sequence of Spd1MPIP. Lower green and blue bars show the Spd11-43 and Spd144-124 deletion mutants. The cyan bar labelled “R1’ represents the location of sequence similarity with a region of Sm11 known to interact with the R1 subunit of RNR [11]. (B) Western blot analysis of Spd1-YFP-NLS levels after HU addition in a wild-type strain (2672), the Spd11-43 mutant (2612), the Spd144-124 mutant (2744) and the Spd1MPIP mutant (2673). Tubulin is shown as a loading control. (C) BiFC of exponentially growing live cells expressing VN173-pcn1 and spd1-VC155 in a wild type (2536) and in a cdt2Δ background (2546). (D) Comparison of the intensity of BiFC in exponentially growing live cells; a wild type (2752) and an spd11-43 (2750) strain (left panels), a wild type (2752) and an spd144-124 (2751) strain (middle panels), and a wild type (2752) and an spd1MPIP (2748) (right panels) strain were compared. Phase (blue) and BiFC signal (green) channels are merged (upper panels). In all the combinations, cells carrying the mutation in Spd1 were stained with MitoTracker® Red to allow direct comparison between the two strains (lower panels). All the strains were in a cdt2Δ background to stabilize Spd1 and comparison of Spd1 levels in these strains is shown in Fig. S3B. (E) Model for RNR activity control by PCNA: left panel represents an unperturbed cell in G1, G2 or M phase that is not subject to DNA damage. Spd1 shuttles between nucleus and cytoplasm, interacting and inhibiting RNR in the cytoplasm and possibly interacting with free PCNA in the nucleus. CRL4Cdt2 does not ubiquitylate nuclear Spd1 as PCNA is not DNA bound. Right panel represents the situation when S phase starts or DNA synthesis associated with DNA damage repair is occurring. Spd1 shuttles from cytoplasm to nucleus but in the nucleus, following interaction with chromatin-bound PCNA, it is ubiquitylated and proteolyzed. The net reduction in Spd1 levels leads to RNR activation.
