Figure 3.
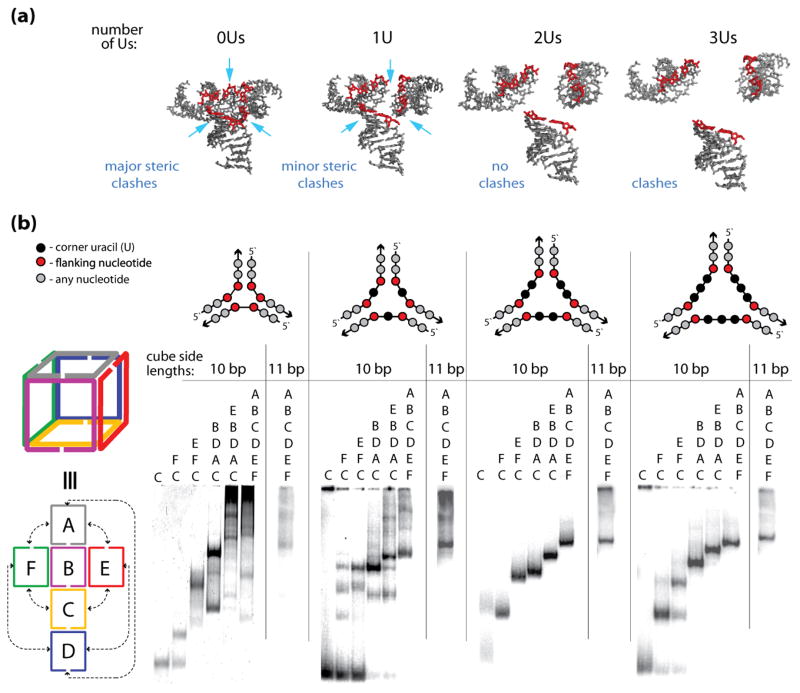
The sizes of the cube corners define the efficiencies of the cube assemblies. (a) 3D models of the cube junctions that were generated to find the minimum number of nucleotides required to link corner helices. The nucleotides indicated in red are the end base pairs that are connected to the single stranded uracil bridges (not shown). The structures correspond to linker loop sizes of 0–3 uracils (Us) respectively. These linker loop sizes are simulated as distance constraints between edge O3′ and P atoms of helices that are designed to be linked. Structure 0U has a distance constraint of maximally 4 Ǻ distance between O3′ and P atoms. This corresponds to no linker loops and the structure has significant steric clashes. A larger distance constraint (1U) of maximally 9 Ǻ corresponds to a structure with 1 residue linker regions. The structural model suggests that the electrostatic interactions between the negatively charged phosphate groups could be somewhat unfavorable and that the cube formation would benefit from the linkers being at least 2 nt-long (structures 2Us and 3Us). (b) native-PAGE experiments indicating the assembly patterns of the RNA nanocubes having zero, one, two, and three single-stranded corner uracils (0U, 1U, 2Us, and 3Us respectively) and corresponding cubes with additional base-pair per each side (11 bp). Color-coded three-dimensional and two-dimensional schematic representations of interactions for the A–F cube sequences (shown to the left of the gel lanes) and schematic representations of cube corners (above the gels) are shown. Corresponding sequence strands are indicated above each lane of the gel.