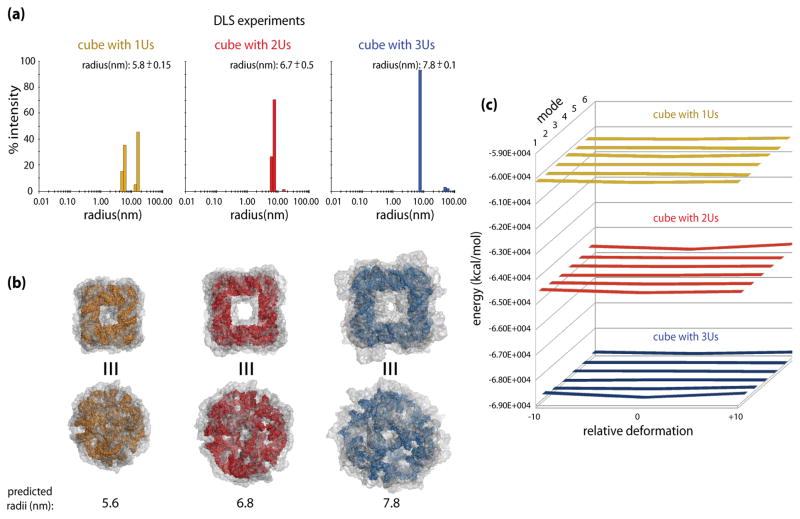
Figure 4.
Predicted and measured dimensions of the nanocubes with 1U, 2U and 3U single-stranded corner linkers, connecting 10bp-long helical edges. (a) Dynamic light scattering (DLS) experiment results for RNA nanocubes having one, two, and three single-stranded corner uracils respectively (1U in yellow, 2Us in red, and 3Us in blue). (b) Colored models of the maximum size cubes (refer to the text for details); dark yellow for the 1U cube, red for the 2U cube, and blue for the 3U cube. Top views show cube faces, while the bottom views focus on one of the truncated corners linked with single strands. The transparent gray envelopes of motion around the cubes illustrate maximum extents of the distortions predicted by the ANM simulations for the top six normal modes. The envelopes delineate the increase in the apparent dimensions of the cubes due to distortions of their initial shapes, which bring them into agreement with the experimentally observed results. (c) The energy plots for the 1U, 2U, and 3U cubes (color-coded as elsewhere) based on the minimized energies of the starting models (relative deformation of 0) and the maximum distortions (−10 and +10). Relative deformations correspond to structural snapshots at discrete points in the calculated distortion trajectories. Data for the top six modes of motion is shown, with mode 1 being the closest to the viewer. The plots show nearly flat (within a fraction of one percent) energy landscapes for the top 6 mode distortions, indicating free mobility of the models to the limits of steric clashes. Refer to the text for details.