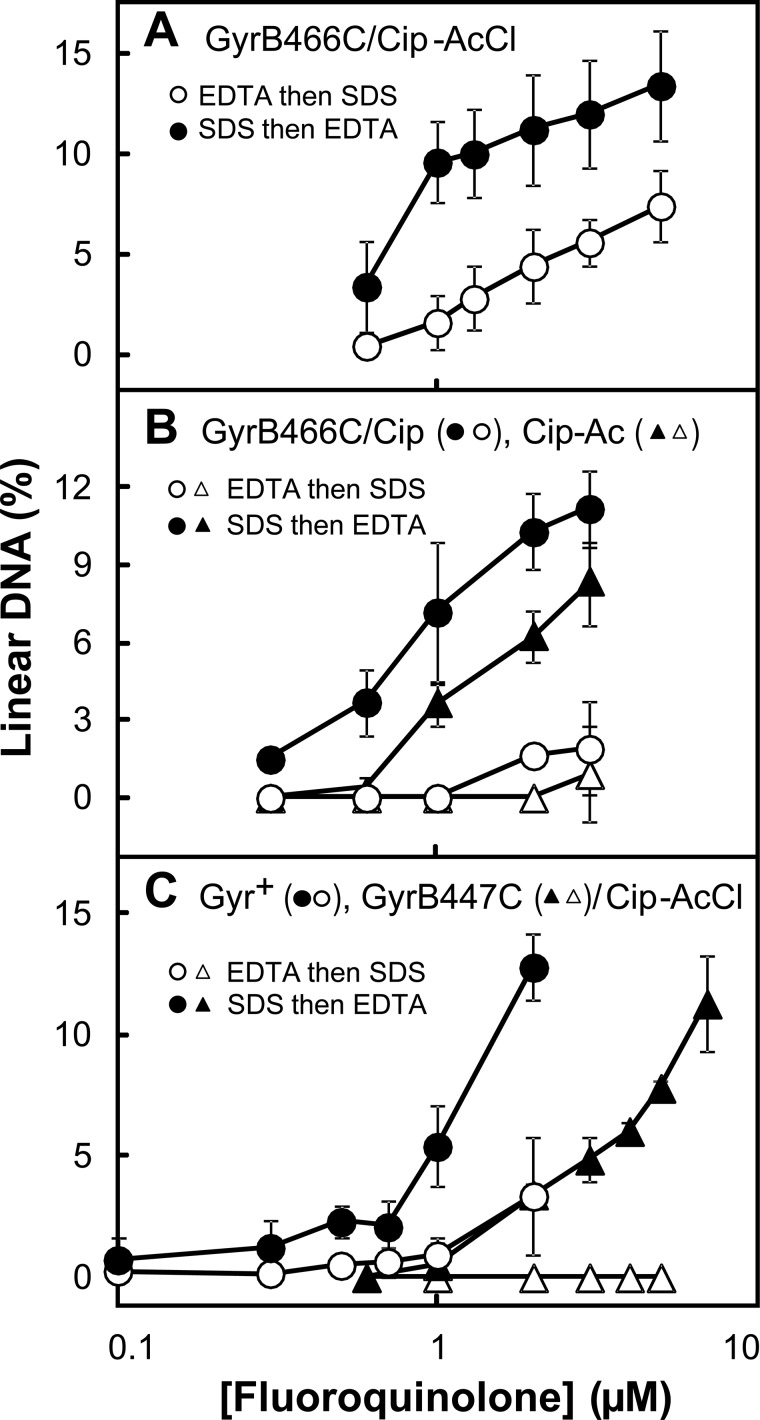
FIGURE 2.
Evidence for cross-linking of GyrB-E466C gyrase to Cip-AcCl. Mixtures containing gyrase, gel-purified supercoiled pBR322 DNA, and ciprofloxacin-based derivatives were incubated at 37 °C. Then reaction mixtures were treated with either SDS and proteinase K (filled symbols) to release DNA breaks generated by drug-gyrase action or with EDTA (empty symbols) to reseal DNA breaks. The SDS-treated samples were then treated with EDTA, and the EDTA samples were treated with SDS and proteinase K before samples were subjected to gel electrophoresis to separate the DNA species. The percentage of linear DNA within each sample was determined as a measure of DNA breaks generated by gyrase-drug action at the indicated drug concentrations. In the data shown, concentrations of gyrase and DNA were 20 and 7.4 nm, respectively; reactions were incubated for 15 min. Error bars indicate S.D. A shows GyrB-E466C gyrase incubated with the cross-linking agent Cip-AcCl; B shows results for GyrB-E466C gyrase with two non-cross-linking agents, ciprofloxacin (circles) and Cip-Ac (triangles); and C shows wild-type gyrase (circles) or GyrB-Cys447 gyrase (triangles) with the cross-linking agent Cip-AcCl. Examination of a variety of enzyme:DNA ratios and incubation times produced results similar to those shown. The number of total experiments and those used for the data shown (chosen so panels had the same conditions), respectively, were 14 and 5 for the comparison of A, 7 and 4 for ciprofloxacin in B, 4 and 4 for Cip-Ac in B, 3 and 3 for GyrB-Cys447 gyrase in C, and 11 and 4 for wild-type gyrase in C.