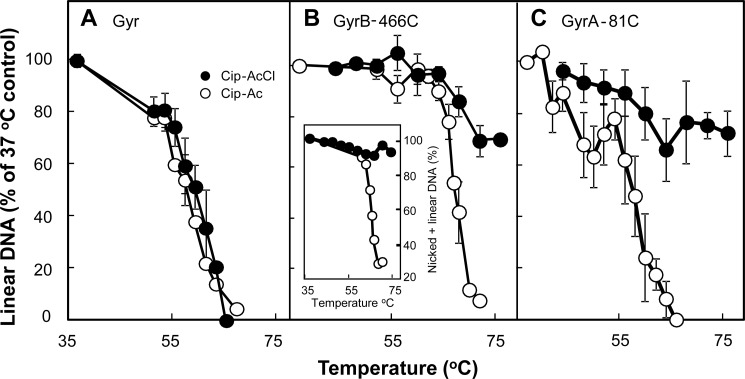
FIGURE 5.
Effect of temperature on DNA resealing. Reaction mixtures containing gyrase, plasmid DNA, and derivatives of ciprofloxacin were incubated for 45 min at 37 °C followed by an additional 5-min incubation at the indicated temperature. Reactions were stopped by chilling on ice; additional SDS-proteinase K treatment and electrophoretic analysis were as described under “Experimental Procedures.” The percentage of DNA in the linear form was determined for each temperature; data were then normalized to the percentage of linear DNA observed at 37 °C to allow comparison of thermal resealing curves containing different levels of linear DNA prior to thermal treatment. A, wild-type gyrase (40 nm) and pBR322 (3.7 nm) were incubated with Cip-Ac (6 μm; empty circles) or Cip-AcCl (6 μm; filled circles). B, GyrB-E466C gyrase (40 nm) and pBR322 DNA (3.7 nm) were incubated with Cip-Ac (80 μm; empty circles) or Cip-AcCl (10 μm; filled circles). The inset shows the effect of thermal incubation on the sum of linear plus nicked DNA for complexes formed with Cip-Ac (empty circles) and Cip-AcCl (filled circles). C, GyrA-G81C gyrase (80 nm) and pBR322 (3.7 nm) were incubated with Cip-Ac (700 μm; empty circles) or Cip-AcCl (500 μm; filled circles). Elevated quinolone concentrations were required to obtain linear DNA at levels similar to those used in A and B (above 30%). Error bars indicate S.D. for the following number of determinations: A, Cip-AcCl, 4; Cip-Ac, 3; B, Cip-AcCl, 4; Cip-Ac, 3; C, Cip-AcCl, 8; Cip-Ac, 5.