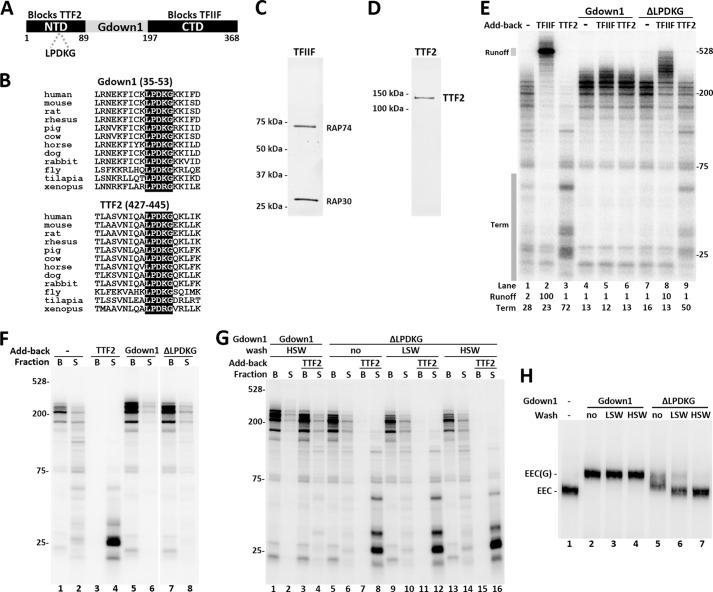
FIGURE 1.
The LPDKG motif in the Gdown1 NTD is required for TTF2 inhibition and tight association with Pol II. A, diagram of Gdown1 indicating the conserved NTD and CTD with their proposed respective functions. The regions that were cloned to make truncation proteins are highlighted and have amino acid numbers marked. Location of the LPDKG motif is indicated. B, protein sequence alignments of Gdown1 and TTF2 in example species around the LPDKG motif. C, silver staining of purified TFIIF (consisting RAP74 and RAP30 subunits). D, silver staining of purified recombinant human TTF2 protein. E, in vitro transcription reactions comparing the effects of Gdown1 (1 pmol/reaction) and the ΔLPDKG mutant (1 pmol/reaction) on TFIIF (0.1 pmol/reaction) and TTF2 (0.04 pmol/reaction) during elongation. Isolated EECs were incubated with indicated factors before a 7-min chase. RNA was isolated and then analyzed on a denaturing gel. The labeled transcripts shown here were detected by phosphorimaging. Transcript sizes are indicated in nucleotides. Intensity of the runoff region (indicated by a shaded box) in each lane was quantified and normalized against lane 2. The termination region (indicated by a shaded box) in each lane was quantified and is shown as its percentage of the total lane intensity. F, in vitro transcription reaction similar to E, except that TTF2 was 0.08 pmol/reaction and after stopping elongation. Beads (B, engaged complexes) and supernatant (S, terminated complexes) were separated and run separately. G, isolated EECs were incubated with 1 pmol of Gdown1 or the ΔLPDKG mutant before being subjected to no wash or two rounds or washing with 60 mm KCl (LSW) or 1.6 m KCl (HSW). The resulting complexes were supplemented with TTF2 (0.08 pmol/reaction) as indicated before a 7-min chase. Beads and supernatant were separated after the chase. H, EC-EMSA showing Gdown1 associating with EECs. Isolated EECs were incubated with 1 pmol of Gdown1 or the ΔLPDKG mutant before LSW or HSW as indicated. The complexes were digested off the beads by SacI at 37 °C for 15 min and run on a 4% native acrylamide gel for 2.5 h at 6 W. Bands corresponding to EEC or EEC(G) are indicated.