FIGURE 2.
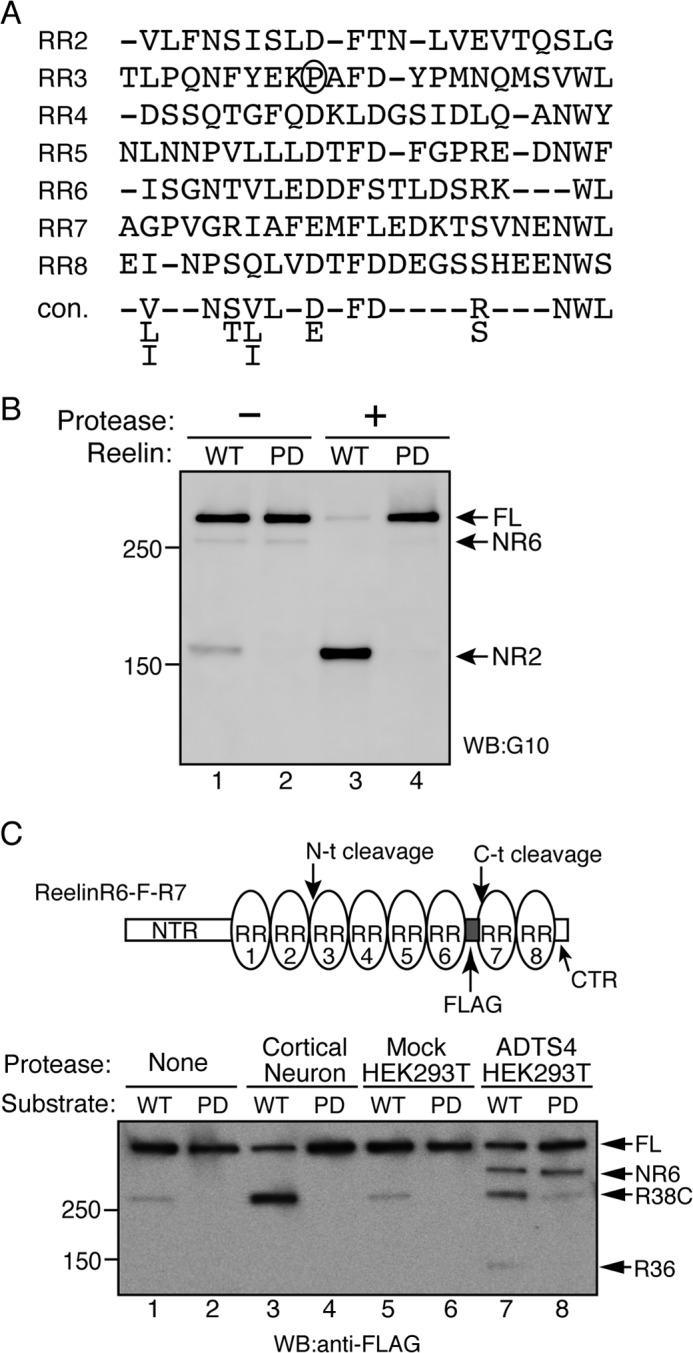
Pro-1244 is necessary for N-t cleavage. A, alignment of the partial primary sequences of the RRs according to Ichihara et al. (22). The consensus sequence (con.) is provided at the bottom of the alignment. Pro-1244 of RR2 is circled. B, Pro-1244 is required for N-t cleavage. Reelin-WT (lanes 1 and 3) or its mutant, Reelin-PD (Pro-1244 substituted with aspartic acid, lanes 2 and 4), were incubated for 24 h with the control buffer (lanes 1 and 2) or partially purified N-t site protease (lanes 3 and 4). The reaction mixtures were analyzed by Western blotting (WB) using anti-Reelin NTR antibody G10. Positions of molecular mass markers (kDa) are shown on the left. FL, full-length. C, N-t cleavage by ADAMTS-4 is inhibited significantly by PD mutation. The schematic of the substrate used in this assay (ReelinR6-F-R7) is shown on the top. ReelinR6-F-R7 (WT, lanes 1, 3, 5, and 7) or its PD mutant (PD, lanes 2, 4, 6, and 8) were incubated with control buffer (None, lanes 1 and 2), the N-t protease partially purified from the culture supernatant of cortical neurons (Cortical Neuron, lanes 3 and 4), the culture supernatant of mock-transfected HEK293T cells (Mock HEK293T, lanes 5 and 6), or that of ADAMTS-4-transfected HEK293T cells (ADTS4 HEK293T, lanes 7 and 8) at 37 °C for 24 h. The reaction mixtures were separated by SDS-PAGE and analyzed by Western blotting with anti-FLAG antibody. Positions of full-length substrate (FL), NR6, R38C, and R36 fragments are indicated by arrows. Positions of molecular mass markers (kDa) are shown at the left.
