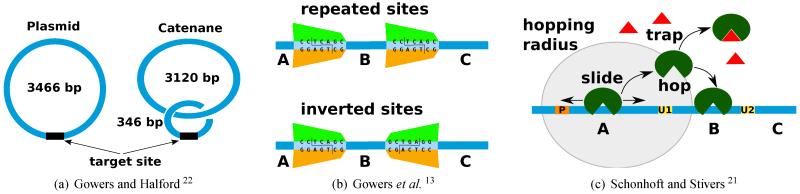
Fig. 2. Experimental strategies aimed to prove the existence of hopping.
(a) The addition of non-specific DNA co-linear with an enzyme restriction site leads to similar cleavage rate as in the case when the non-specific DNA is added by catenation22. In the second scenario the restriction site is reachable from the catenane only if hopping exists. (b) The experimental setup assumes two DNAs each with two restriction sites, but while in the first DNA, the sites are repeated (and no reorientation of the enzyme is required), in the second DNA, the sites are inverted (and the enzyme needs to invert its orientation which is possible only through hopping). Ensuring that only one enzyme is bound to the DNA, Gowers et al. 13 observed that for distances longer than 50 bp the two strands display similar cleavage rates at both sites. The processivity of the two sites is measured as P = ([A] + [C] − [BC] − [AC])/([A] + [C] + [BC] + [AC]). (c) This is a similar strategy as in (b), but the experiments assumes only one DNA with two damaged uracil sites where the hUNG enzyme can excise the DNA. The protein is released from a P site and if the protein leaves the DNA for long excursions then it gets inactivated by a trap molecule and, thus, only a pure sliding mechanism will ensure a similar excision rate as in the case of a system without the trap molecule.