Fig. 3.
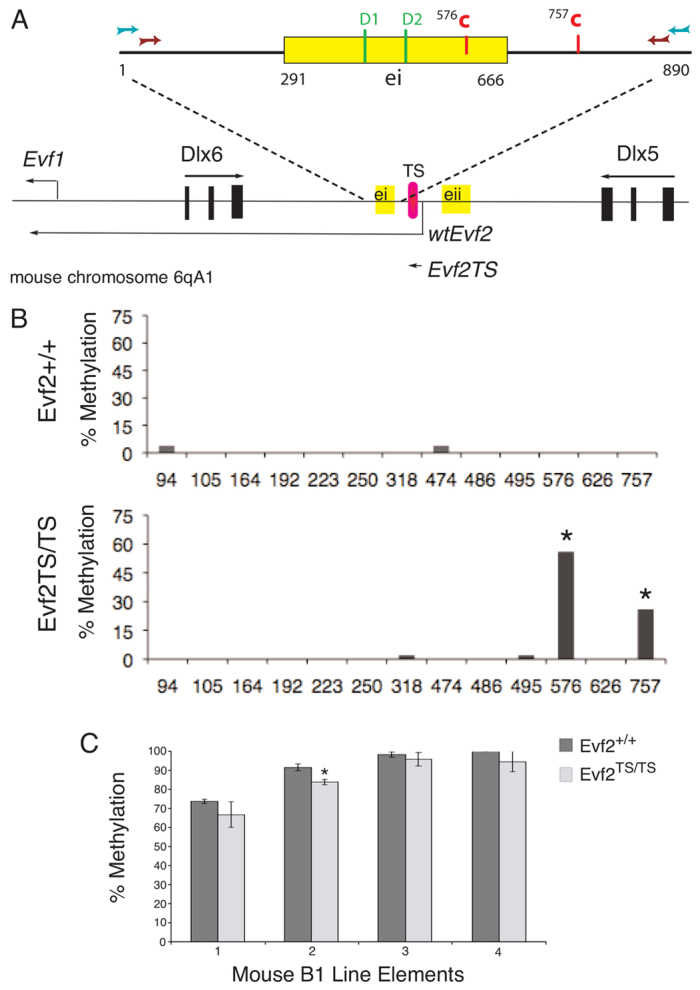
Evf2 lncRNA prevents site-specific CpG DNA methylation within the Dlx5/6 ultraconserved enhancer ei. E13.5 MGE DNA isolated from three Evf2+/+ and three Evf2TS/TS mutants was bisulfite treated, PCR amplified, subcloned, and individual clones sequenced. (A) Schematic of the Dlx5/6 intergenic region, containing the intergenic enhancers ei (ultraconserved) and eii, with expansion of the 890 PCR region spanning ei (blue and brown arrows indicate nested primers, where blue arrows indicate external primers, and brown arrows indicate internal primers). There are 13 possible CpG DNA methylation sites within this 890-nucleotide (nt) region. 576CpG and 757CpG are each marked by a red C. Pink oval represents the location of the triple poly(A) transcription stop (TS) insertion site at the 5′ end of Evf2 in Evf2TS/TS mice. The wild-type Evf2 transcript is ∼3.7 kb, whereas Evf2TS generates a predicted truncated transcript (80 nt) before transcription termination. DLX1/2 binding sites, as previously identified (Zerucha et al., 2000), within ei are in green (D1 and D2). (B) Graph of percentage methylation comparing Evf2+/+ and Evf2TS/TS E13.5 MGE at 13 possible CpG sites within the 890 PCR region shown in A. Data are obtained from 52 Evf2+/+ and 56 Evf2TS/TS individual clones. n=3 for each genotype. Loss of Evf2 results in increased methylation at sites 576CpG and 757CpG, *P<0.01. (C) Global methylation analysis of four different B1 line elements (1-4) in Evf2+/+ and Evf2TS/TS E13.5 MGE DNA shows that Evf2 loss does not increase global methylation. Error bars represent s.e.m.
