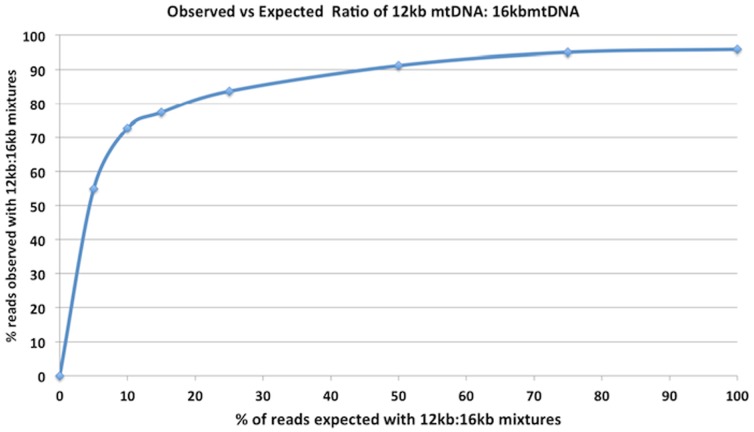
Figure 2. Deleted Mitochondrial DNA hyper-amplifies with LR-PCR.
Observed vs Expected coverage of two unique haplogroup mtDNA samples pooled prior to LR-PCR amplification. One 4.5 kb Kearns-Sayre homozygous deleted mtDNA (12.1 kb, KSS mtDNA) sample is mixed with a known wild type mtDNA (16.6 kb, NA12878 mtDNA) sample with a different haplogroup. The KSS mtDNA sample has a unique haplogroup that creates heteroplasmies at expected loci when mixed with a full length mtDNA control. After sequencing the mixtures to 10,000× mean coverage on an Illumina MiSeq V2 system, allele frequencies are measured across a barcoded dilution series where the deleted sample alleles are expected to be seen at 5%,10%,15%,25%,50%,75% of the reads. Plotted is the expected coverage of the KSS mtDNA alleles versus the observed ratio (Y-Axis) of the control mtDNA alleles. This is measured by mapping reads with Bowtie and counting allele frequencies at the haplogroup specific loci. This result is expected in that a multiplexed PCR containing 12.1 kb and16.6 kb molecules will selectively amplify the smaller template. The selective amplification was still observed despite 15 minute extension times applied in PCR. This also highlights the pronounced sensitivity for detecting large deletions in mtDNA samples using LR-PCR.