Figure 2.
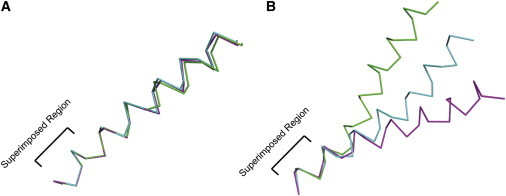
Comparison of helical structures restrained by absolute (orientational) restraints (A) and relative (distance) restraints (B). Calculations were performed in XPLOR-NIH. An extended conformation was equilibrated at 3500 K for 20 ps using torsion angle restraints (φ = −60°, ψ = −45°, ±30°) followed by slow cooling simulated annealing from 3500 to 100 K. Both calculations used torsion angle, helical hydrogen bond, bond angle, improper dihedral, and van der Waals restraints for simulated annealing. The absolute calculation was based on an ideal helix tilted at 25° that was also restrained by 20 15N chemical shift anisotropy restraints using a generous error bar of ±10 ppm and tensor element values of δ11 = 57.3, δ22 = 81.2, δ33 = 227.8ppm (2), as well as 20 1H-15N dipolar coupling restraints with an error bar of ±0.5 kHz and dipolar magnitude of 10.375 kHz (2,3). The relative calculation was also based on the same ideal helix and was also restrained by 20 nitrogen (i, i + 4) and 20 α-carbon (i, i + 4) distance restraints (5Å cutoff). All restraints were modeled as square-well potentials and the scaling of the force constants for the structure calculation was the same as that used for the Influenza A M2 monomer (4). Three representative, lowest energy, structures are shown for each calculation. Superposition of the structures was calculated using the backbone atoms of residues 1–6 of each helix.
