Abstract
The American College of Medical Genetics and Genomics (ACMG) recently released guidelines regarding the reporting of incidental findings in sequencing data. Given the availability of Direct to Consumer (DTC) genetic testing and the falling cost of whole exome and genome sequencing, individuals will increasingly have the opportunity to analyze their own genomic data. We have developed a web-based tool, PATH-SCAN, which annotates individual genomes and exomes for ClinVar designated pathogenic variants found within the genes from the ACMG guidelines. Because mutations in these genes predispose individuals to conditions with actionable outcomes, our tool will allow individuals or researchers to identify potential risk variants in order to consult physicians or genetic counselors for further evaluation. Moreover, our tool allows individuals to anonymously submit their pathogenic burden, so that we can crowd source the collection of quantitative information regarding the frequency of these variants. We tested our tool on 1092 publicly available genomes from the 1000 Genomes project, 163 genomes from the Personal Genome Project, and 15 genomes from a clinical genome sequencing research project. Excluding the most commonly seen variant in 1000 Genomes, about 20% of all genomes analyzed had a ClinVar designated pathogenic variant that required further evaluation.
1. Background and Significance
The era of personalized genomics received a jumpstart in 2007, when 23andMe, deCODEme, and Navigenics began to offer Direct to Consumer (DTC) personal genetic testing.1 Reports from these companies include genotyping of up to hundreds of thousands of loci with phenotypic interpretation for dozens to hundreds of traits and conditions based mainly upon genome wide association studies (GWAS).2,3 The use of such genetic information in a clinical setting has been slower to develop, although several academic medical centers have established genomic medicine programs.4 Moreover, with the falling price of next generation sequencing, the number of whole genomes and exomes being sequenced is steadily increasing.4,5 Whole genome or exome sequencing provides much more data than genotyping, especially with regards to rare and private mutations. As a consequence, incidental findings in an individual’s genome beyond the scope of the research or clinical question are likely to exist. There is some debate surrounding the handling of the so-called “incidentalome”, particularly since novel, rare, or private mutations may be difficult to interpret and a full interpretation is cost prohibitive in most settings.6 Recently, the American College of Medical Genetic and Genomics (ACMG) put out a report with recommendations on which incidental findings should be specifically analyzed and reported.7 In this case, “incidental findings” refer to pathogenic or potentially pathogenic variants discovered in a subset of genes during whole genome or exome sequencing, regardless of the reason sequencing was ordered.7,8 The list of 57 genes covering 24 conditions put forward by the ACMG are those that have medically actionable outcomes. For example, the list includes BRCA1 and TNNI3, mutations in which can lead to breast cancer and hypertrophic cardiomyopathy, respectively.7 Currently, it is not known exactlywhat percentage of individual genomes will carry such variants, and an understanding of the pathogenic burden will allow researchers to better understand the resources required to evaluate such variants. Here, we present a publicly available tool, PATH-SCAN, which annotates genomes for ClinVar designated pathogenic variants in the list of genes recommended by the ACMG.7
2. Methods
PATH-SCAN allows a researcher or individual to analyze and annotate individual exomes or genomes for a set of pathogenic variants identified in the ClinVar database in the genes put forward by the ACMG. These annotations are presented in a report with genomic information and links to additional information. Due to the consequences of many of these variants, security and privacy are mainstays of the PATH-SCAN program. PATH-SCAN maintains complete privacy by performing all analyses on an individual’s local machine, similar to a previously described genotype analysis tool, INTERPRETOME.9 PATH-SCAN offers an option to anonymously submit data to our research group allowing us to use crowd sourcing to determine the prevalence of pathogenic variants found in the ACMG gene list.
2.1.Pathogenic Variant Selection
Pathogenic variants were selected from the National Center for Biotechnology Information’s (NCBI) ClinVar variant call file (VCF). From this database of variants, those variants with at least one submission as “pathogenic” were extracted and annotated with links to other clinically relevant databases. Since the ClinVar database is a collaborative database with potentially variable quality in individual variant results, we filtered out any variant that was tagged with a “variant suspect” code. A variant might be labeled as such for several reasons, including being called from an old genomic alignment or a suspected paralog From this list, we then extracted only the variants that mapped to the 57 genes listed in the ACMG report. Gene boundaries were determined using GRCh37.p10.10 In total, ClinVar had records for 994 variants designated as pathogenic across 57 genes. These variants are included in the PATH-SCAN package. The original ClinVar VCF can be found here http://www.ncbi.nlm.nih.gov/variation/docs/human_variation_vcf/. The use of other databases is allowed in case an individual wishes to use an alternative database for annotation (see Appendix).
2.2.Analysis Tool
Our cross-platform program, PATH-SCAN, utilizes a database of 994 variants to scan personal genomes and annotate them. The annotations produced by PATH-SCAN are made available to the end user or researcher as a local html page with a simplified user interface for increased accessibility and transparency. To assist interpretation of this information and provide a model for future genome interpretation tools, each recognized variant and annotation is presented alongside links to relevant educational resources, including ClinVar, OMIM, and consolidated Gene Reviews from the National Center for Biotechnology Information (NCBI).
Crowd sourcing data collection was accomplished by making a submission link available that transfers de-identified anonymous information back to our data collection server. In order to prevent any privacy concerns regarding this data collection, PATH-SCAN only transmits the total number of pathogenic variants annotated for each gene (e.g. the total pathogenic burden per gene of an individual genome) as well as a unique key to prevent duplicate submissions from unwary users. Additional information such as ancestry is optional to transmit. The unique key is calculated by PATH-SCAN automatically by hashing the personal genome file using the SHA-2 family of cryptographic functions. In addition to these security measures, a privacy message is presented before the user can submit their data. For the personal genomes we had direct access to, the full annotations made by PATH-SCAN were used to collect data on individual diseases and variants as well as aggregate distributions of pathogenic variants across individuals.
PATH-SCAN is a command line utility that was developed in Python 2.7.5 and has no external dependencies. The PATH-SCAN program comes pre-loaded with the existing database of pathogenic variants. We also have the ability to load updated databases pending re-releases of the ACMG recommendation or for custom made variant databases. PATH-SCAN will automatically detect and process variant call files (VCFs), tab-separated variant (TSV) files from Complete Genomics, and SNP chip results from 23andMe. Because 23andMe only genotypes SNPs, PATH-SCAN will not scan data in this form for indels. For a whole genome VCF file that is 336 MB, PATH-SCAN runs in 24 seconds on a machine with 16GB of RAM and a 2.3 ghz processer. The script and database are bundled and available for download online at: http://montgomerylab.stanford.edu/pathscan.zip.
2.3.Applying PATH-SCAN to existing datasets: 1000 Genomes, Personal Genomes Project, and a clinical sequencing project
We pilot tested PATH-SCAN on the 1092 individuals from the 1000 Genomes project publicly available low coverage (~4x) genomes.11 We also investigated how ancestry affected the number of variants found in each population. Additionally, we tested PATH-SCAN on exome chip data for 2123 individuals from the 1000 Genomes project. These individuals overlap with the 1092 whole genome data.11
We also tested our tool on 163 Genomes downloaded from the Personal Genomes Project, which were in the Complete Genomics format (www.personalgenomes.org/community.html).12 We only considered variants called with high quality. High quality variants are called on homozygous calls with a quality score greater than or equal to 20 and heterozygous calls with a quality score greater than or equal to 40 under the maximum likelihood variable allele fraction.
In addition to the larger scale, low-coverage studies previously discussed, we tested our tool on a clinical sequencing project consisting of 15 individuals (3 trios and 4 unrelated individuals).
3. Results
3.1.Pathogenic variants studied
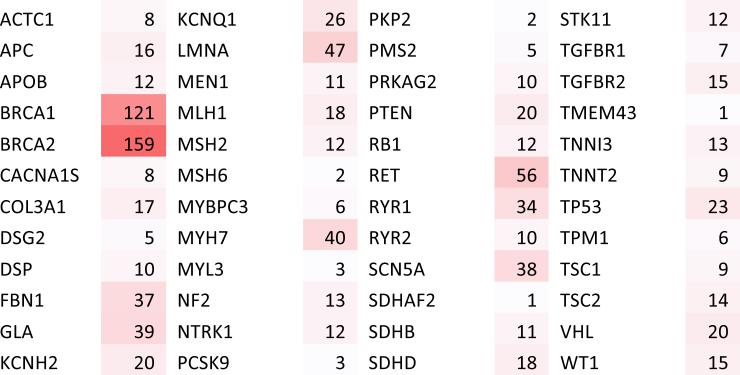
By filtering ClinVar for variants with evidence of pathogenicity in the subset of ACMG guideline genes, we selected 994 variants that our tool evaluates. These variants include 651 single nucleotide polymorphisms (SNPs) and 343 small insertions/deletions (indels). 65.5% of the pathogenic variants evaluated were SNPs, evenly distributed across all 12 non-synonymous nucleotide-to-nucleotide transversions. Variants were not evenly distributed across the 57 genes, with BRCA1 and BRCA2 having the largest number of variants (Figure 1). An example of the output of PATH-SCAN can be seen in Figure 2.
Figure 1.
Total number of pathogenic variants found per gene in ClinVar. In total there were 994 variants distributed across the 57 genes specified by the ACMG recommendations.
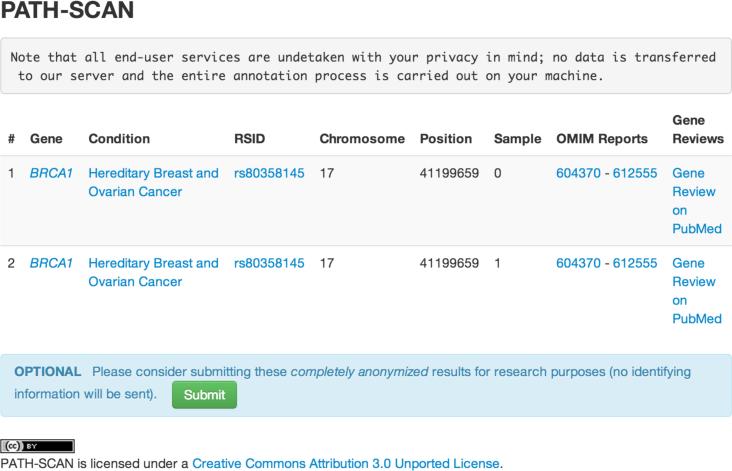
Figure 2.
Sample output of PATH-SCAN. Information regarding the affected variant (including chromosome, position, rsID, and gene) are displayed alongside relevant information including what condition this variant is expected to have pathology in and links to clinical reviews and publications regarding the condition. A crowd-sourcing form is available at the bottom of the page if users wish to submit de-identified information to our servers.
3.2.PATH-SCAN identifies variants in 1000 Genomes Data
Out of 1092 individuals with low coverage genome data, 633 have at least one ClinVar designated pathogenic variant reported in one of the ACMG genes. Out of the 2123 exome-chipped individuals (which overlaps with the 1092 individuals with whole genomes), 997 individuals had at least one variant reported. The most common variant seen was rs1805124 (SCN5A), which was seen in 41.2% of individuals (Table 1). This variant has an allele frequency of about 20% in the 1000 Genomes population. Excluding this very common variant, out of 1092 low coverage genomes, 225 individuals had at least one pathogenic variant in one of the ACMG genes, and 237 individuals had at least one pathogenic variant in the exome chip data.
Table 1.
Variants and individual frequencies seen in the 1000 Genomes Project Data. Absent data from the exome chip columns due to incomplete sequencing coverage in those individuals. Frequencies represent frequency of individuals with at least one copy of the variant and not allele frequencies.
| Gene | Disease | rsID | 4× Genome (1,092 indv.) | Freq. | Exome Chip (2,123 indv.) | Freq. |
|---|---|---|---|---|---|---|
| APC | Familial adenomatous polyposis | rs137854567 | 2 | 0.002 | - | - |
| rs1801166 | 8 | 0.007 | - | - | ||
| DSP | Arrhythmogenic right-ventricular cardiomyopathy | rs121912998 | 4 | 0.004 | - | - |
| LMNA | Hypertrophic cardiomyopathy, dilated cardiomyopathy | rs57830985 | 1 | 0.001 | - | - |
| MSH6 | Lynch syndrome | rs2020912 | 11 | 0.010 | 13 | 0.006 |
| SCN5A | Romano–Ward long QT syndrome types 1, 2, and 3, Brugada syndrome | rs1805124 | 450 | 0.412 | 852 | 0.401 |
| rs41261344 | 26 | 0.024 | 72 | 0.034 | ||
| rs45620037 | 1 | 0.001 | - | - | ||
| rs7626962 | 26 | 0.024 | 65 | 0.031 | ||
| SDHB | Hereditary paraganglioma–pheochromocytoma syndrome | rs11203289 | 19 | 0.017 | - | - |
| rs33927012 | 17 | 0.016 | 30 | 0.014 | ||
| SDHD | Hereditary paraganglioma–pheochromocytoma syndrome | rs11214077 | 20 | 0.018 | - | - |
| rs34677591 | 13 | 0.012 | - | - | ||
| STK11 | Peutz–Jeghers syndrome | rs59912467 | 28 | 0.026 | 61 | 0.029 |
| TP53 | Li–Fraumeni syndrome | rs28934576 | 1 | 0.001 | - | - |
| TSC1 | Tuberous sclerosis complex | rs118203576 | 48 | 0.044 | - | - |
| rs118203657 | 5 | 0.005 | - | - |
3.3.PATH-SCAN identifies variants in the Personal Genomes Project
We applied PATH-SCAN to 163 genomes in Complete Genomics format. 77 of these individuals were found to have at least one variant. The most common variant, once again, was rs1805124 (Table 2). Excluding this variant, 27 individuals had at least one variant in one of the ACMG guidelines genes.
Table 2.
Variants and counts seen in 163 Personal Genomes
| Gene | Disease | rsID | PGP Genomes (163 individuals) |
|---|---|---|---|
| APC | Familial adenomatous polyposis | rs1801166 | 5 |
| DSG2 | Arrhythmogenic right-ventricular cardiomyopathy | rs193922639 | 2 |
| FBN1 | Marfan syndrome, Loeys–Dietz syndromes, and familial thoracic aortic aneurysms and dissections | rs137854475 | 1 |
| KCNQ1 | Romano–Ward long QT syndrome types 1, 2, and 3, Brugada syndrome | rs267607197 | 1 |
| RET | Multiple endocrine neoplasia type 2; Familial medullary thyroid cancer | rs77724903 | 1 |
| SCN5A | Romano–Ward long QT syndrome types 1, 2, and 3, Brugada syndrome | rs1805124 | 62 |
| rs41261344 | 1 | ||
| rs137854610 | 1 | ||
| SDHB | Hereditary paraganglioma–pheochromocytoma syndrome | rs33927012 | 7 |
| SDHD | Hereditary paraganglioma–pheochromocytoma syndrome | rs11214077 | 5 |
| rs34677591 | 1 | ||
| STK11 | Peutz-Jeghers syndrome | rs59912467 | 1 |
| TNNT2 | Hypertrophic cardiomyopathy, dilated cardiomyopathy | rs121964857 | 1 |
| TSC1 | Tuberous sclerosis complex | rs118203657 | 1 |
3.4.Analyzing variant burden across populations
We looked at the variant detection in the different 1000 Genomes populations (Table 3). Because of the high allele frequency of rs180524, we looked at the frequencies with and without this SNP.
Table 3.
Number of variants seen in the different 1000 Genomes populations.
| Population | 4× Genome Samples (1092 total) | Avg. variant count/ person 4× Genome | Avg. variant count/person 4× Genome w/o rs180524 | Exome Chip Samples (2123 total) | Avg. variant count/person Exome Chip | Avg. variant count/person Exome Chip w/o rs180524 |
|---|---|---|---|---|---|---|
| ACB | 0 | - | - | 98 | 71/0.72 | 20/0.20 |
| ASW | 61 | 44/0.72 | 12/0.20 | 97 | 63/0.65 | 10/0.10 |
| CDX | 0 | - | - | 100 | 36/0.36 | 24/0.24 |
| CEU | 85 | 45/0.53 | 13/0.15 | 104 | 41/0.39 | 5/0.05 |
| CHB | 97 | 44/0.45 | 20/0.21 | 100 | 44/0.44 | 20/0.2 |
| CHD | 0 | - | - | 1 | 0/0 | 0/0 |
| CHS | 100 | 35/0.35 | 21/0.21 | 150 | 44/0.37 | 35/0.23 |
| CLM | 60 | 47/0.78 | 21/0.35 | 107 | 52/0.46 | 2/0.19 |
| FIN | 93 | 45/0.48 | 13/0.14 | 100 | 40/0.4 | 4/0.04 |
| GBR | 89 | 53/0.60 | 13/0.15 | 101 | 54/0.53 | 9/0.09 |
| GIH | 0 | - | - | 93 | 42/0.45 | 4/0.04 |
| IBS | 14 | 18/1.29 | 4/0.29 | 147 | 87/0.59 | 12/0.08 |
| JPT | 89 | 35/0.39 | 10/0.11 | 100 | 37/0.37 | 10/0.1 |
| KHV | 0 | - | - | 118 | 56/0.47 | 38/0.32 |
| LWK | 97 | 64/0.66 | 9/0.09 | 100 | 64/0.64 | 6/0.06 |
| MKK | 0 | - | - | 31 | 22/0.71 | 1/0.03 |
| MXL | 66 | 47/0.71 | 26/0.39 | 100 | 35/0.35 | 5/0.05 |
| PEL | 0 | - | - | 104 | 46/0.44 | 0/0 |
| PUR | 55 | 49/0.89 | 21/0.38 | 111 | 62/0.56 | 3/0.02 |
| TSI | 98 | 69/0.70 | 22/0.22 | 100 | 57/0.57 | 9/0.09 |
| YRI | 88 | 85/0.97 | 25/0.28 | 161 | 129/0.80 | 24/0.15 |
ACB- African Caribbean in Barbados; ASW - HapMap African ancestry individuals from Southwest US; CDX- Chinese Dai in Xishuangbanna, China; CEU – Utah residents with Northern and Western European ancestry; CHB - Han Chinese in Beijing; CHD - Chinese in metropolitan Denver, CO; CHS – Southern Han Chinese; CLM - Colombian in Medellin, Colombia; FIN -HapMap Finnish individuals from Finland; GBR - British individuals from England and Scotland; GIH - HapMap Gujarati India individuals from Texas; IBS - Iberian populations in Spain; JPT – Japanese in Tokyo, Japan; KHV - Kinh in Ho Chi Minh City, Vietnam; LWK - Luhya individuals in Webuye, Kenya; MKK- HapMap Maasai individuals from Kenya; MXL - HapMap Mexican individuals from LA California; PEL - Peruvian in Lima, Peru; PUR- Puerto Rican in Puerto Rico; TSI – Tuscans from Italy; YRI- Yoruba from Ibadan, Nigeria
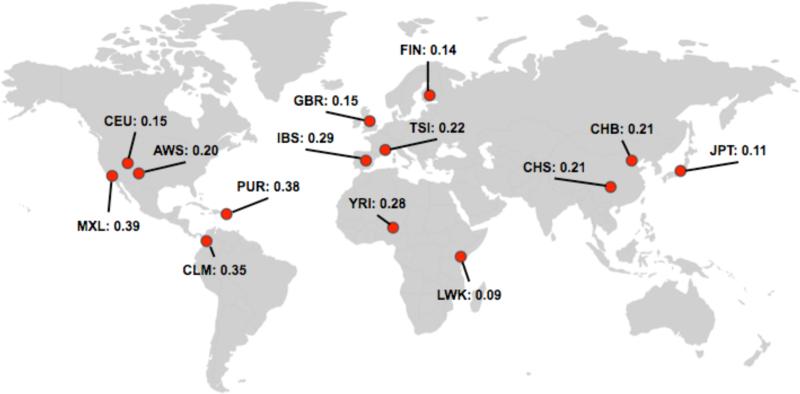
In 1092 Genomes, the average number of variants per genome ranged from 0.35 (CHS) to 1.29 (IBS). Without rs180524, the average number of variants per person ranged from 0.09 (LWK) to 0.39 (MXL). Populations that were closely related had similar average variants per person (Figure 3). Particular populations, such as LWK, had a much lower variant count than other populations when rs180524 was not taken into consideration.
Figure 3.
Average variants per individual in 1092 Genomes (with rs1805124 removed due to high allele frequency in all populations).
3.5.Applying PATH-SCAN to a clinical genome sequencing project
In a clinical genome sequencing project consisting of 15 individuals, 2 subjects had 2 ClinVar pathogenic variants, 5 subjects had 2 ClinVar pathogenic variants, and 8 subjects had 0 ClinVar pathogenic variants. The variant list was not directly reported to us due to IRB constraints.
4. Discussion
Since the ACMG report on incidental findings was published, there has been much debate around explicitly searching for and reporting variants in the ACMG’s gene list.8,13 Issues have included the difficulty of substantiating which variants are pathogenic, the cost of additional screening, and the lack of information about how often variants are seen and how many each individual could possibly carry. Here, we present a tool, which serves as an example of how technicians, researchers, clinicians, and individuals may screen for potentially pathogenic and actionable variants. Furthermore, we have applied this tool on existing datasets and have made it available for public use in order to gauge the frequency that potentially pathogenic variants in the ACMG genes are observed.
4.1.Variant Selection
One of the major issues was outlined in the original ACMG report: “The Working Group recognized that there is no single database currently available that represents an accurately curated compendium of known pathogenic variants, nor is there an automated algorithm to identify all novel variants meeting criteria for pathogenicity.”7 For the purposes of this project, we selected the ClinVar database, because the variants submitted come directly from patient data. We selected only those variants that had at least one submission indicating that the variant was pathogenic in nature. A limitation of this approach is the inclusion of variants that may have conflicting submissions listing the variant as pathogenic and benign, and issues such as sample size and study population can contribute to this confusion about variant interpretation. However, the ClinVar curators are making an effort to review submissions. We recognize that variants labeled as pathogenic by ClinVar may not be viewed as so when analyzed by a clinical laboratory, genetic counselor, or clinician. However, their presence in a genome or exome will warrant evaluation in order to determine if they should be acted upon. Thus, understanding the frequency of such variants will allow us to draw conclusions about the amount of resources required to properly vet variants in the ACMG guidelines genes.
Another limitation of our database choice is that we do not pick up novel, rare, or private mutations that are not currently annotated in ClinVar. However, since we could not reliably make any inference about the pathogenicity of such variants, we selected not to include them in our publicly available tool. Finally, because most research studies are done in individuals of European descent, there is likely an overrepresentation of variants that are pathogenic in populations of European descent.14
We do note that the pathogenic variants in the ClinVar database are not evenly distributed between genes. The number of pathogenic variants reported in a gene can be influenced by several factors – including the length of the gene, the amount of selective pressure, and the number of studies focusing on the gene. Interestingly, BRCA1 and BRCA2 had the largest number of pathogenic variants. This could be due to the extensive studies on these genes and their role in hereditary breast and ovarian cancer.
4.2.Findings in the 1000 Genomes Data and Personal Genomes Project
Our successful application of PATH-SCAN to the 1000 Genomes data sets confirmed the ability of our tool to process whole genomes. In 1092 low pass genomes, 566 individuals had a pathogenic variant in one of the ACMG genes.
The most observed variant was rs1805124 (H558R), seen in 41.2% of individuals. The population allele frequency of this variant is about 20% in 1000 Genomes. This is a prime example of the challenge with implementing an automatic system to follow up on potentially pathogenic variants in ACMG genes. SCN5A H558R has been associated with atrial fibrillation and changes in cardiac conduction.15,16 Multiple studies have also demonstrated that the presence of this variant combined with other rare SCN5A variants perturbs heart electrophysiology.17–19 However, there are also studies in which this variant may mitigate the effects of a particular mutation that causes Brugada syndrome.20 Finally, it should be noted that this variant is quite common in the general population. As Klitzman et al. noted in response to the ACMG Guidelines, ‘pathogenic’ variants with a high frequency in the population but a low corresponding disease prevalence may cause unnecessary alarm.13 Because this variant can affect disease risk when other mutations are also present, its presence would require evaluation of the entire gene and family history by an experienced genetic counselor or clinician. This example supports the need for comprehensive follow up of variants that are thought to be pathogenic.
Excluding rs1805124 (H558R), 233 individuals out of 1092 carried an incidental finding. These other variants were less common, with less than 5% of individuals carrying any single variant. These variants included risks for such conditions as colon cancer (rs1801166) and cardiomyopathy (rs121912998), which can profoundly impact health and lifestyle. 21,22
When we looked across the populations, we saw that there were differences in the average number of variants per person. Because many of these variants were derived from studies done in individuals of European ancestry, differences could be attributed to this selection bias.14 Furthermore, different populations likely have different variants driving their total variant counts due to differences in population allele frequency. In the case of LWK, which had a very low average variant per person count when the most common variant was removed, we are likely missing population specific pathogenic variants. Another complex issue brought up by ancestry is pathogenicity – variants that may be causative and pathogenic in one population may not have the same penetrance or impact in another.14 With our crowdsourcing tool, ancestry will be an option that individuals can submit; we hope that this will allow us to get a more accurate picture of the distribution of these variants across individuals of different and mixed ancestries.
We also note that since these are low coverage genomes (~4x), some variants reported could be false. Genomes sequenced to clinical standards would have much higher coverage and have more confident calls. Thus, this data may be skewed by false positives.
To evaluate our tool on Complete Genomics data and higher coverage genomes, we applied PATH-SCAN to 163 genomes made publicly available from the Personal Genomes Project. Once again, rs1805124 (H558R) was the most common variant. However, excluding this variant, 17% of genomes had variants of interest. Overrepresentation of certain variants may occur if individuals in the Personal Genome Project are related. Several of these variants were low frequency at a population level, as they did not appear in the 1000 Genomes data. Our tool assists in the evaluation of such variants by pinpointing them within minutes of scanning a genome.
4.3.Using PATH-SCAN on Clinical Genomes
Finally, we ran PATH-SCAN on a clinical genome sequencing cohort of fifteen individuals. The output provided a starting point for the evaluation of variants in the project. Previously, people used a gene-based approach to look at all variants in a gene of interest and then used manual curation to select variants for further evaluation.
4.4.PATH-SCAN as a quantitative evaluation tool
PATH-SCAN is a publicly available tool; individuals using it can choose to anonymously submit their pathogenic burden (i.e. the number of variants seen in their genome) and ancestry to our server. Over time, we aim to use crowdsourcing to get a more accurate number of how often potentially pathogenic variants are seen and how ancestry affects these numbers.
The current iteration of our tool serves as the foundation for additional functionalities in development. Because ClinVar designated pathogenic variants may not truly be pathogenic, we are currently working on adding variant effect prediction scores, such as PolyPhen and SIFT to our tool.23,24
We have found that even with the most common pathogenic variant removed, a substantial percentage of individuals still carry variants in ACMG guidelines genes that require additional investigation. Of course, due to the limitations of the ClinVar database, many of these variants may be benign. However, we feel that each variant needs to be evaluated in the context of other mutations, clinical history, and family history by a clinician or genetic counselor. While not all of these variants may be ultimately reported back, evaluating these variants will require additional resources. Thus, understanding how often such variants occur is key to assessing the resource utilization of following the ACMG Guidelines. In the past few months, there has much debate surrounding the ACMG Guidelines and their implementation. Our tool PATH-SCAN aims to streamline the identification of variants in ACMG recommended genes that warrant further investigation and to provide data on how often each variant is seen.
Acknowledgments
RBA and TEK are funded by NIH/NIGMS NIGMS R24 GM61374. SBM is funded by the Edward Mallinckrodt Jr. Foundation. RD is funded by Stanford MSTP and T32 HG000044. ZZ is funded by NSF GRFP and T32 HG000044. KEO is funded by 5 P50 HG003389-05
6. Appendix A
PATH-SCAN Manual Download http://montgomerylab.stanford.edu/pathscan.zip
Requirements: Python 2.7.5; a web browser
Command Line Interface A full description of the CLI for PATH-SCAN follows:
$ python pathscan.py <genome file> [--suppress | --db <database>]
<genome file> is either a VCF file, a Complete Genomics TSV file, or a 23andMe SNP file.
--suppress If this flag is specificed PATH-SCAN will only report data on the command line.
--db <database file> Can be used to specify a different database file. The database format is a TAB-delimited file with 9 columns, all required. First column is chromosome, second is position, third is RSID, fourth is the reference allele, fifth is the alternate allele, sixth is the gene name, seven is the gene review ID numbers (can be replaced with a '.'), eight is the OMIM ID number (can be replaced with a '.', and the ninth is the clinical significance code from ClinVar (can be replaced with a '.').
References
- 1.Gurwitz D, Bregman-Eschet Y. Personal genomics services: whose genomes? European journal of human genetics : EJHG. 2009;17:883–9. doi: 10.1038/ejhg.2008.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kalf RRJ, et al. Variations in predicted risks in personal genome testing for common complex diseases. Genetics in medicine : official journal of the American College of Medical Genetics. 2013:1–7. doi: 10.1038/gim.2013.80. doi:10.1038/gim.2013.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vernez SL, Salari K, Ormond KE, Lee SS-J. Personal genome testing in medical education: student experiences with genotyping in the classroom. Genome medicine. 2013;5:24. doi: 10.1186/gm428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Manolio T. a, et al. Implementing genomic medicine in the clinic: the future is here. Genetics in medicine: official journal of the American College of Medical Genetics. 2013;15:258–67. doi: 10.1038/gim.2012.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Metzker ML. Sequencing technologies - the next generation. Nature reviews. Genetics. 2010;11:31–46. doi: 10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]
- 6.Kohane I, Masys D, Altman R. The incidentalome: a threat to genomic medicine. JAMA: the journal of the. 2006;296:212–215. doi: 10.1001/jama.296.2.212. [DOI] [PubMed] [Google Scholar]
- 7.Green RC, et al. ACMG recommendations for reporting of incidental findings in clinical exome and genome sequencing. Genetics in medicine: official journal of the American College of Medical Genetics. 2013 doi: 10.1038/gim.2013.73. doi:10.1038/gim.2013.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Allyse M, Michie M. Not-so-incidental findings: the ACMG recommendations on the reporting of incidental findings in clinical whole genome and whole exome sequencing. Trends in biotechnology. 2013;31:439–441. doi: 10.1016/j.tibtech.2013.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karczewski K, Tirrell R. Interpretome: A freely available, modular, and secure personal genome interpretation engine. Pac. Symp. 2012 [PMC free article] [PubMed] [Google Scholar]
- 10.Consortium IHGS. Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–45. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- 11.Abecasis GR, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lunshof JE, Chadwick R, Vorhaus DB, Church GM. From genetic privacy to open consent. Nature reviews. Genetics. 2008;9:406–11. doi: 10.1038/nrg2360. [DOI] [PubMed] [Google Scholar]
- 13.Klitzman R, Appelbaum PS, Chung W. Return of Secondary Genomic Findings vs Patient Autonomy Implications for Medical Care. JAMA. 2013;310:369–370. doi: 10.1001/jama.2013.41709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rosenberg N. a, et al. Genome-wide association studies in diverse populations. Nature reviews. Genetics. 2010;11:356–66. doi: 10.1038/nrg2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen L, et al. Polymorphism H558R in the Human Cardiac Sodium Channel SCN5A Gene is Associated with Atrial Fibrillation. Journal of International Medical Research. 2011;39:1908–1916. doi: 10.1177/147323001103900535. [DOI] [PubMed] [Google Scholar]
- 16.Gouas L, et al. Association of KCNQ1, KCNE1, KCNH2 and SCN5A polymorphisms with QTc interval length in a healthy population. European journal of human genetics: EJHG. 2005;13:1213–22. doi: 10.1038/sj.ejhg.5201489. [DOI] [PubMed] [Google Scholar]
- 17.Cheng J, et al. SCN5A rare variants in familial dilated cardiomyopathy decrease peak sodium current depending on the common polymorphism H558R and common splice variant Q1077del. Clinical and translational science. 2010;3:287–94. doi: 10.1111/j.1752-8062.2010.00249.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Makielski JC, et al. A ubiquitous splice variant and a common polymorphism affect heterologous expression of recombinant human SCN5A heart sodium channels. Circulation research. 2003;93:821–8. doi: 10.1161/01.RES.0000096652.14509.96. [DOI] [PubMed] [Google Scholar]
- 19.Ye B, Valdivia CR, Ackerman MJ, Makielski JC. A common human SCN5A polymorphism modifies expression of an arrhythmia causing mutation. Physiological genomics. 2003;12:187–93. doi: 10.1152/physiolgenomics.00117.2002. [DOI] [PubMed] [Google Scholar]
- 20.Poelzing S, et al. SCN5A polymorphism restores trafficking of a Brugada syndrome mutation on a separate gene. Circulation. 2006;114:368–76. doi: 10.1161/CIRCULATIONAHA.105.601294. [DOI] [PubMed] [Google Scholar]
- 21.Frayling I, Beck N. The APC variants I1307K and E1317Q are associated with colorectal tumors, but not always with a family history. Proceedings of the. 1998 doi: 10.1073/pnas.95.18.10722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang Z, et al. Desmosomal dysfunction due to mutations in desmoplakin causes arrhythmogenic right ventricular dysplasia/cardiomyopathy. Circulation research. 2006;99:646–55. doi: 10.1161/01.RES.0000241482.19382.c6. [DOI] [PubMed] [Google Scholar]
- 23.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nature protocols. 2009;4:1073–81. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 24.Adzhubei I. a, et al. A method and server for predicting damaging missense mutations. Nature methods. 2010;7:248–9. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]