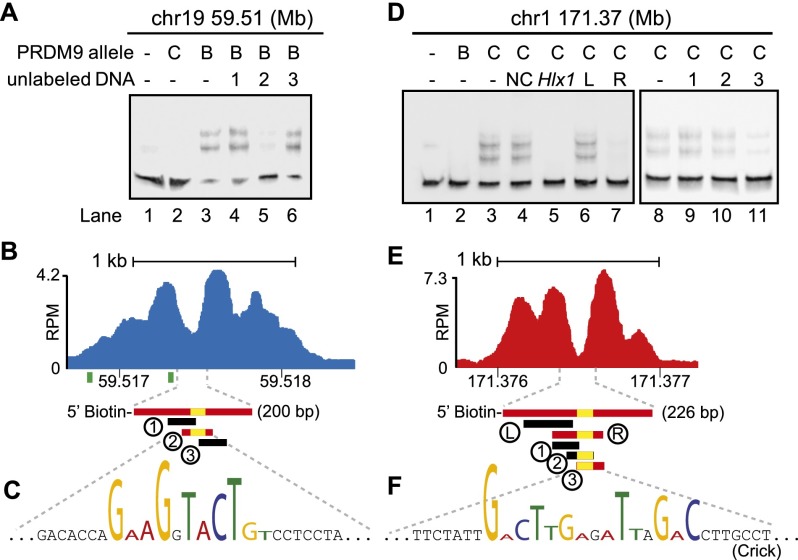
Figure 4.
PRDM9 binds hotspots at the NDR. (A) Mapping of a PRDM9Dom2 binding site. EMSA shows specific binding of PRDM9Dom2 at the PRDM9Dom2-activated hotspot located on chromosome 19 at Mb 59.51. Only unlabeled DNA oligo 2, containing the consensus motif, can compete for binding (Lane 5). All lanes contain labeled DNA; otherwise, the composition of the binding reactions is shown above the blot (B: PRDM9Dom2; C: PRDM9Cst). (B) ChIP-seq coverage profile from Prdm9Dom2 showing H3K4me3 nucleosome positions, the NDR, and DNA sequences used for the EMSA in A (red bars indicate DNA that binds PRDM9Dom2; black bars indicate nonbinding sequences; yellow bars show the position of the PRDM9Dom2 motif; green squares under the graph show the position of the predicted binding site from Getun et al. [2012]). (C) The partial DNA sequence of oligo 2. Nucleotides matching the PRDM9Dom2 consensus motif in Figure 2E are formatted to indicate similarity. (D–F) Similar to A–C except showing results for PRDM9Cst. (D) PRDM9Cst binds hotspot chr1 171.36 Mb at the NDR ([NC] Noncompeting oligo 2 from chr19 59.51 in panel A. [Hlx1] Binding site for Hlx1 [chr1 186.44 Mb] from Billings et al. [2013]). (E) ChIP-seq coverage profile at hotspot chr1 171.37 from B6-Prdm9CAST-KI. (F) Partial DNA sequence of oligo 3 indicating nucleotides that match the consensus in Figure 2F.