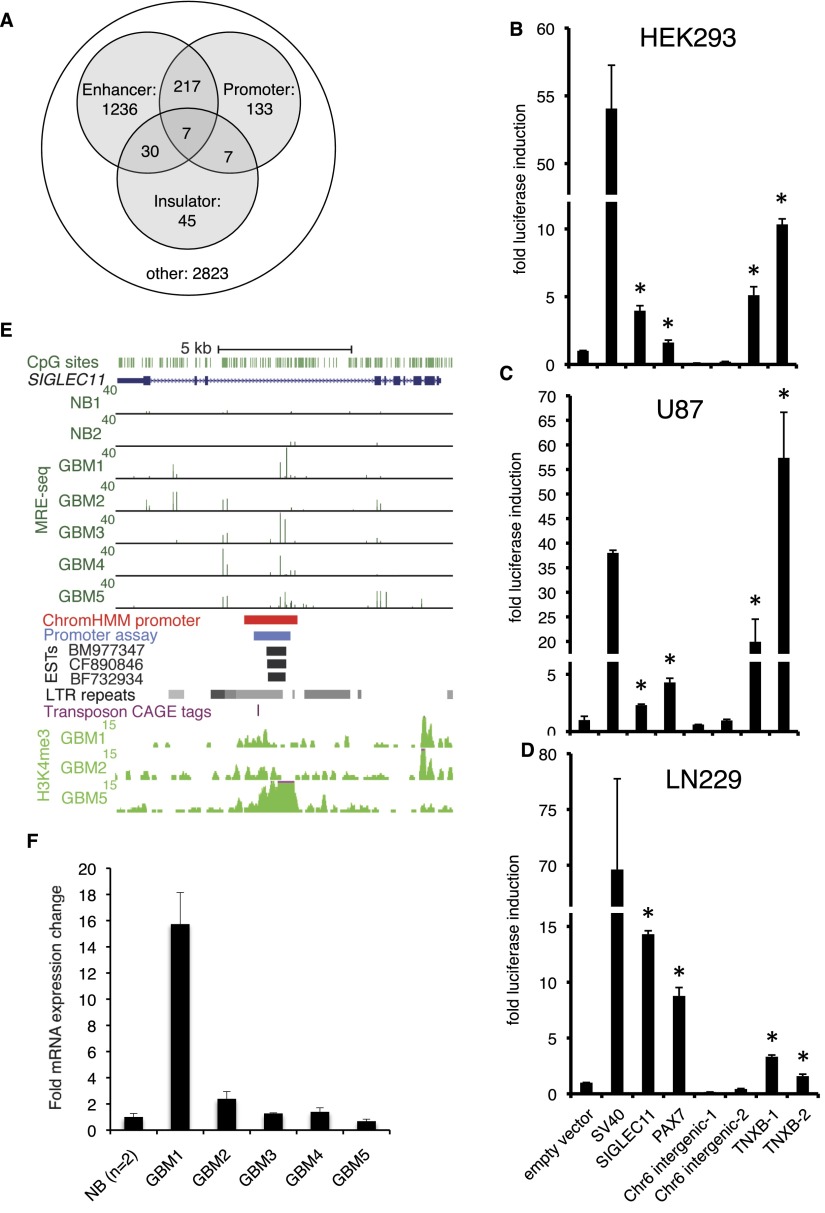
Figure 3.
Recurrent DNA hypomethylation of previously unannotated promoters in GBM. (A) ChromHMM-defined promoters, enhancers, and insulators from nine ENCODE cell lines overlap with recurrent GBM hypomethylated DMRs (Q < 10−5 in at least two of five GBMs). (B–D) Promoter assays for DNA sequences within hypomethylated DMRs that were not associated with the 5′ ends of RefSeq or UCSC transcripts, but had a promoter state assigned by ChromHMM in at least one ENCODE cell type. Each bar shows fold induction (±SD) relative to pGL3-Basic empty vector control. pGL3-SV40 promoter is used as a positive control, shown immediately to the right of the empty vector. These candidate promoters were tested in HEK293 cells and two GBM cell lines, U87 and LN229. Asterisks indicate P < 0.05, one-tailed t-test. (E) Previously unannotated alternative promoter in the body of SIGLEC11 showing GBM hypomethylation and overlapping promoter state by ChromHMM. The region cloned for luciferase assay is shown in blue, and human ESTs suggesting transcription initiation are shown below (black). LTR repeats from UCSC RepeatMasker track, including the HERV3 element in which the promoter is embedded, are indicated with gray. The location of a transposon-associated CAGE tag cluster (Faulkner et al. 2009) is shown by the purple hatch mark. H3K4me3 ChIP-seq signal in GBMs 1, 2, and 5 is shown at the bottom. (F) qRT-PCR with primers located within human EST BM977347 (see E). Fold expression ± SD relative to normal brain is graphed for a representative experiment. (NB) Normal brain.