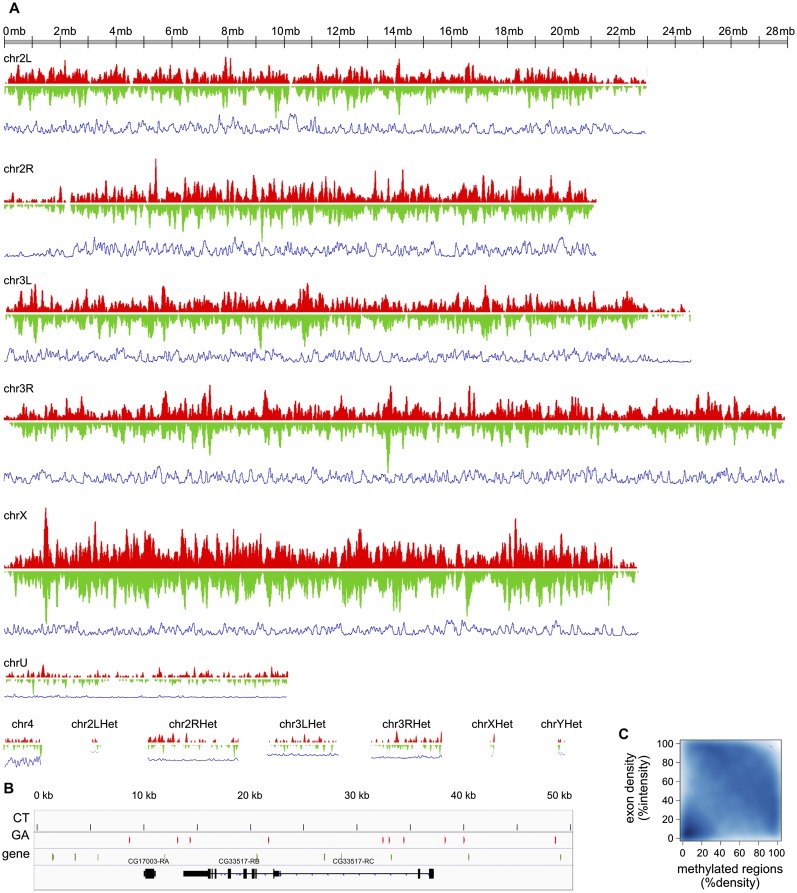
Figure 2.
Genomewide distribution of methylation. (A) For both CT (red) and GA (green) strands, the density of methylated regions was calculated with Fseq using a feature length of 100 kb. Density of methylated regions on the CT and GA strands is plotted on the vertical axis with the chromosome displayed horizontally. The blue lines below each chromosome represent exon density calculated in the same way. The scale of the vertical axis is the same on all the plots. This reveals that density of methylation is highest on the X chromosome and lowest on the heterochromatic chromosomes. The density on the CT and GA strands is roughly symmetrical at this large scale. (B) Detailed example of the strand-asymmetric distribution of methylated regions along a 50-kb genomic interval containing two annotated genes. Regions methylated on the CT strand are shown in red, and regions methylated on the GA strand are in green. (C) Anticorrelation between the density of methylated regions and the density of exons over 100-kb genomic intervals. The scatterplot displays the density distribution of methylated regions over 100-kb intervals on the x-axis, and the exon density distribution over the same size intervals on the y-axis . The intensities of the distributions of methylated regions and exons are percentile-normalized. The bottom left corner shows that many regions of low exon density are nearly devoid of methylation. The concentration visible across the upper right half indicates that regions of high exon density tend to have low methylation density, and regions of high methylation density tend to have low exon density.