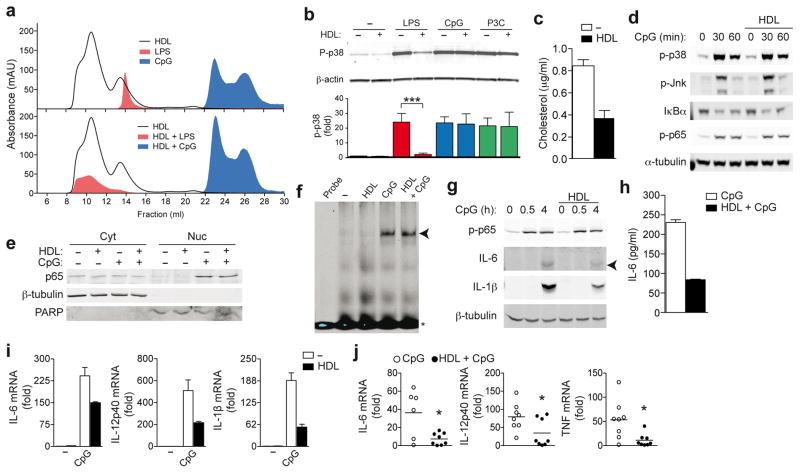
Figure 2.
HDL inhibits TLR-induced pro-inflammatory cytokine transcription. a, Bodipy-labeled LPS, Alexa 647-labeled CpG 1826 or HDL were run over an S200 size exclusion column separately (a, upper panel) or together (a, lower panel) and absorbance profiles examined. b, LPS (200 ng/ml), CpG (100 nM) or P3C (50 ng/ml) were incubated with HDL (2 mg/ml), and BMDMs stimulated for 30 min. Whole cell lysates were analyzed for p38 phosphorylation (p-p38) relative to total β-actin. c,d BMDMs were pre-treated with HDL (2 mg/ml) for 6 h before stimulation with CpG (100 nM) for indicated times: total cellular cholesterol (c) and phosphorylation of p38, JNK, NF-κB p65 and IκBα degradation (d). e,f BMDMs were pre-treated with HDL as before, and stimulated with CpG (100 nM) for 30 min: subcellular localization of NF-κB p65 (β-tubulin: cytoplasmic loading control; poly ADP ribose polymerase (PARP): nuclear loading control) (e) and NF-κB binding to a target probe as analysed by EMSA (f). g,h BMDMs were pre-treated with HDL as before: CpG-induced phospho-NF-κB p65 and intracellular IL-6 and IL-1β were measured in whole cell extracts, (g); secreted IL-6 measured by ELISA (h). mRNA expression of BMDMs pre-treated with HDL as before, and stimulated with 100 nM CpG for 4 h (i). j, Liver mRNA profile 1 h after C57BL/6 mice were injected with CpG following 6 h HDL pre-treatment (n=8). a, Data is representative of at least three independent experiments. b, Representative immunoblot and densitometric analysis combined from three independent experiments (mean ±S.E.M, each ligand; no HDL versus HDL incubation ***p<0.005). c–i, Representative data from at least three independent experiments (mean ±S.D). j, Mean values ±S.E.M, CpG versus HDL+CpG *p<0.05.