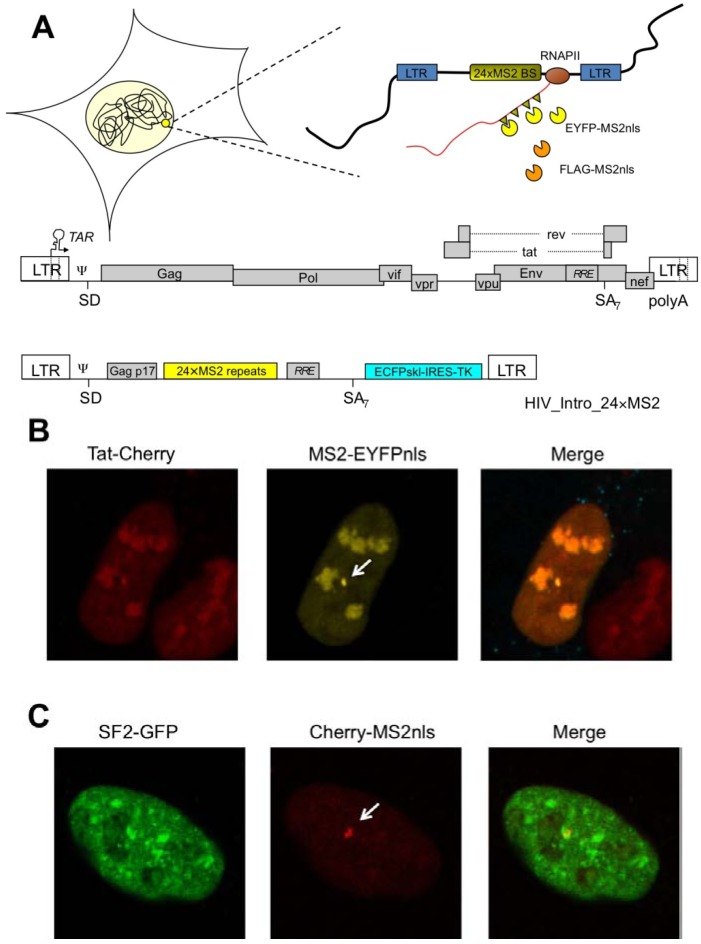
Figure 1.
(a) Schematic description of the HIV-1 MS2-tagging method. Top, the viral cassette carrying the MS2 repeats is integrated in the cell’s chromatin. RNAPII transcribes along the provirus and produces tagged RNAs that are bound by EYFP-MS2nls for visualization of the nascent RNA. Alternatively, FLAG-MS2nls can be used to pull-down the viral RNA for affinity purification of the associated proteome. An outline of the full-lengthviral genome is also shown below with the construct HIV_intro_24xMS2 (not drawn to scale). These constructs are described in great detail in a series of papers [30,32,35]; (b) Localization of Tat on HIV-1 RNA at the transcription site. Tat-Cherry was found associated with the transcription site marked by the accumulation of MS2 (white arrow) in U2OS cell clones expressing EYFP-MS2nls; (c) Localization of SF2 on HIV-1 RNA at the transcription site. SF2-GFP was found associated with the transcription site marked by the accumulation of MS2 (white arrow) in U2OS cell clones expressing Tat and EYFP-MS2nls.