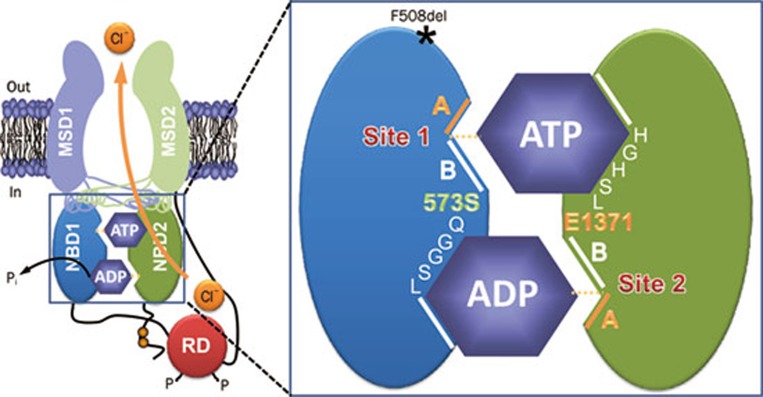
Figure 2.
The organization of the ATP-binding sites in CFTR. The simplified model shows the molecular architecture of ATP-binding site 1 (site 1) and ATP-binding site 2 (site 2) in an open CFTR Cl− channel. Each ATP-binding site is formed by the Walker A and B motifs (labeled A and B, respectively) of one NBD and the LSGGQ motif of the other NBD. Site 2 contains a canonical LSGGQ motif, whereas site 1 contains a non-canonical LSGGQ motif (LSHGH). Site 2 also contains a catalytic base (E1371) at the distal end of the Walker B motif, but this residue is absent in site 1 (S573). The location of the CF mutation F508del on the surface of NBD1 opposite intracellular loop 4 (ICL4) is shown by an asterisk. Abbreviations: MSD, membrane-spanning domain; NBD, nucleotide-binding domain; P, phosphorylation of the RD; Pi, inorganic phosphate; RD, regulatory domain. In and out denote the intra- and extracellular sides of the membrane, respectively. See text for further information. Modified, with permission, from Hwang and Sheppard36.